Figure 6.
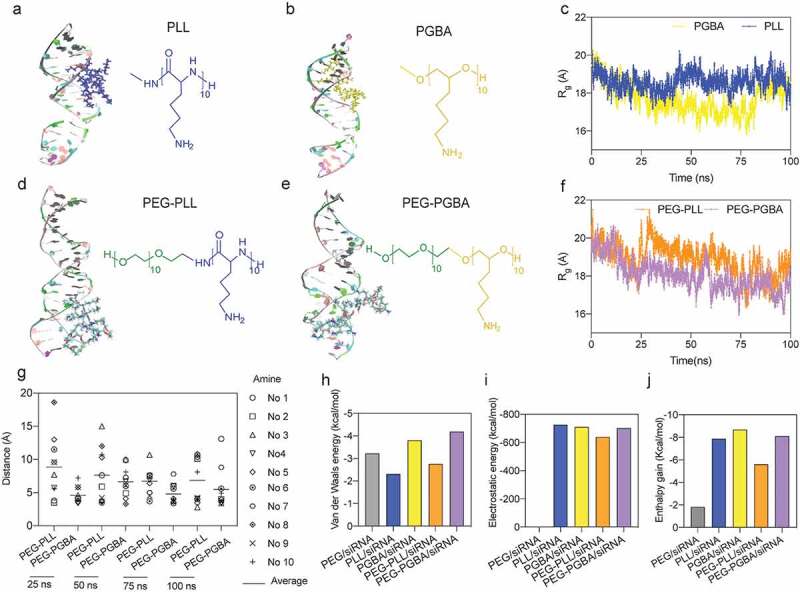
Molecular dynamics (MD) simulation of siRNA binding with polycations and block catiomers. (a), (b) Front view snapshots taken from the dynamic simulation of PLL-siRNA and PGBA-siRNA. (c) Time evolution of Radius of gyration (Rg) of PLL-siRNA and PGBA-siRNA. (d), (e) Front view snapshots taken from the dynamic simulation of PEG-PLL-siRNA and PEG-PGBA-siRNA. (f) Time evolution of Rg of PEG-PLL-siRNA and PEG-PGBA-siRNA. (g) Amine/phosphate distance in PEG-PLL-siRNA and PEG-PGBA-siRNA, each amine is identified by residue numbers (No.). (h) Van der Waals energy, (i) electrostatic energy and (j) enthalpy gain during polymers/siRNA interaction (normalized per charged amine, expressed in kcal mol–1)
