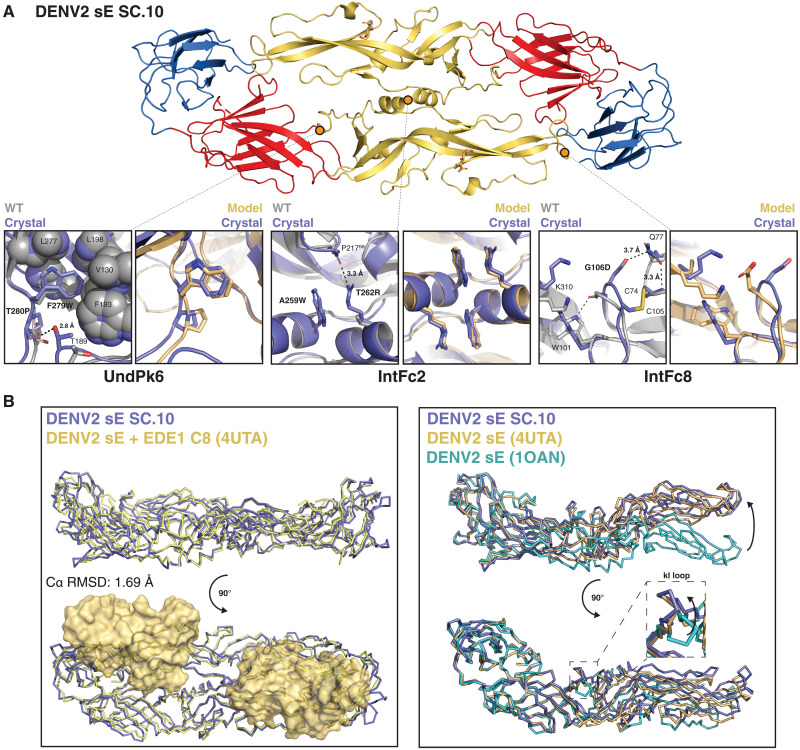
Fig. 5. DENV2 sE SC.10 crystal structure comparison to Rosetta model and EDE1 C8/sE cocrystal structure.
(A) Top: DENV2 sE SC.10 dimer conformation observed in the crystal structure, colored by domain (DI, red; DII, yellow; DII, blue). Bottom: Comparison of UndPk6 (left), IntFc2 (middle), and IntFc8 (right) mutations observed in the SC.10 crystal structure (purple) to DENV2 sE WT crystal structure (gray; PDB 1OAN) and the Rosetta-predicted computational model (yellow). Hydrogen bonds are shown as black dotted lines. (B) Left: Structure superposition of the DENV2 sE SC.10 dimer (purple) with the DENV2 sE WT dimer (yellow) cocrystallized with EDE1 C8, a DENV broadly neutralizing quaternary epitope antibody. EDE1 C8 Fabs are shown as a surface representation. Right: Structure superposition of the DI and DIII domains from DENV2 sE WT sE (PDB 1OAN, teal; and 4UTA, yellow) and DENV2 sE SC.10 (purple). Inset shows a closer view of the kl loop conformation observed in each structure after DI and DIII alignment.