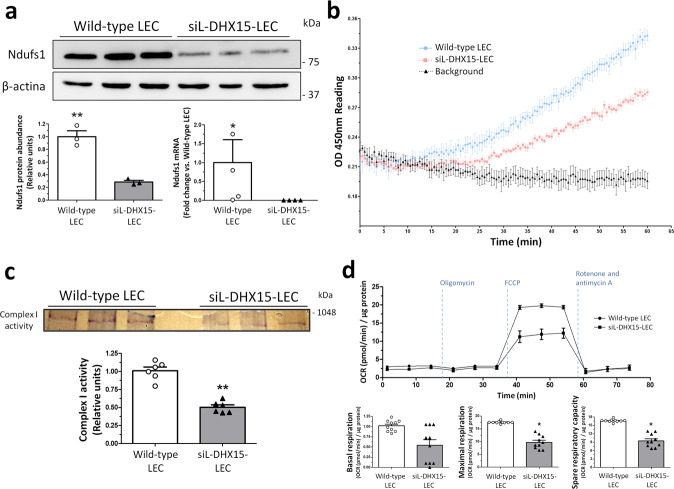
Fig. 5. siL-DHX15-LEC showed impaired mitochondrial respiratory chain activity.
a Protein and gene expression of NDUFS1 was analyzed. Protein was evaluated by western blot using cell lysates from wild-type and siL-DHX15-LEC. β-actin was used as a loading control. The densitometric analysis of the protein expression are shown on the bottom left bar graph. **p < 0.01 (n = 3 biologically independent samples for each condition). mRNA expression was analyzed by RT-qPCR. mRNA levels are illustrated as fold change relative to HPRT mRNA levels. Bottom right graph. *p < 0.05 (n = 4 biologically independent samples for each condition). b Complex I activity was assessed using a commercial colorimetric enzymatic reaction reagent for the mitochondrial respiratory complex I in wild-type and siL-DHX15-LEC. Seven hundred µg of total protein was used in each experimental condition (n = 3 biologically independent samples for each condition). Wild-type-LEC: upper line, siL-DHX15-LEC: middle line, background: bottom line. c In-gel activity staining on clear-native page (CN-PAGE) of the respiratory complex I from wild-type and siL-DHX15-LEC’s mitochondria. The quantification of the relative band intensities of complex I activity is shown in the graph below. **p < 0.01 vs. wild-type LEC (n = 6 biologically independent samples for each condition). d Wild-type and siL-DHX15-LEC were evaluated for basal, coupled and maximal respiration in a mito-stress assay using a Seahorse XFe24 analyzer (n = 10 independent Seahorse wells for each condition). OCR is normalized to µg of total protein. *p < 0.05 vs. wild-type LEC. All statistical analyses were performed using unpaired two-tailed Student’s t-test. All data are presented as mean ± SEM.