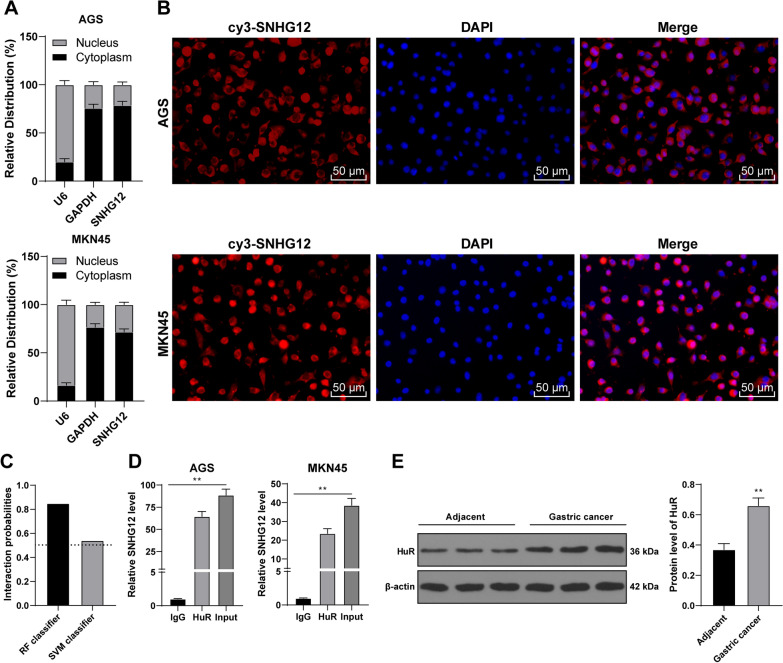
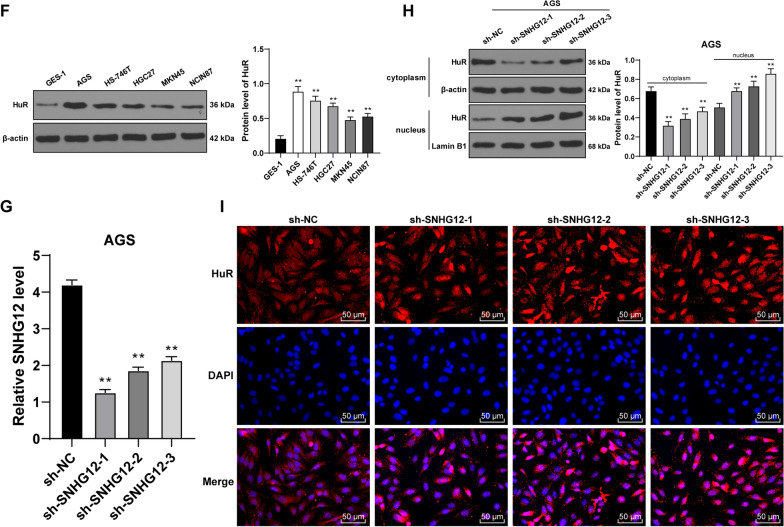
Fig. 6.
SNHG12 bound to RNA-binding protein HuR and induced transportation of HuR from the nuclei to the cytoplasm. A, B The location of SNHG12 was analyzed using subcellular fraction assay and RNA FISH; C The binding probability between SBHG12 and HuR was predicted using RNA–Protein Interaction Prediction (RPISeq) database. D The binding between SNHG12 and HuR was analyzed using RIP assay. E, F The protein expression of HuR in GC tissues and cells was detected by western blot. After the three designed SNHG12 shRNA were treated with AGS cells, NC shRNA was used as the control. G The expression in SNHG12 in AGS cells was detected by RT-qPCR. H The protein expression of HuR in GC cells was detected by western blot. I The aggregation of HuR in the cytoplasm was detected by immunofluorescence. The experiment was repeated three times independently. Data were presented as mean ± standard deviation. Comparison between data in panel E was analyzed by using the t-test. Comparison among groups in panels D, F, G, H was analyzed using one-way ANOVA, followed by Tukey's multiple comparisons test. *p < 0.05, **p < 0.01. sh-NC: NC shRNA; sh-SNHG12: SNHG12 shRNA