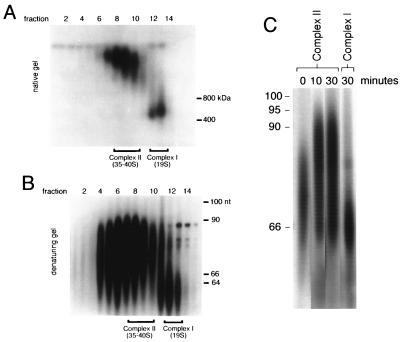
FIG. 2.
Posttranscriptional labeling of isolated mitochondria. Isolated mitochondria were pulse-labeled with [α32P]UTP under conditions that arrest transcription but support polyuridylation of gRNAs. Even numbers at the top of both panels A and B represent glycerol gradient fractions. (A) Native-gel separation of pulse-labeled mitochondrial RNPs fractionated on a 10 to 30% glycerol gradient. Fractions containing complexes I and II are indicated. Protein molecular size markers are indicated in kilodaltons. (B) Analysis of labeled gRNAs present in complexes I and II from the posttranscriptional pulse-labeling experiments on denaturing sequencing gel. Fractions containing complexes I and II are indicated. nt, nucleotides. (C) Time course of pulse-chase-labeled complex II gRNAs shown in panel B. Isolated mitochondria were pulsed with [α32P]UTP for 3 min and then incubated with excess unlabeled UTP for the times shown (for details, see Materials and Methods). DNA size standards are indicated in nucleotides.