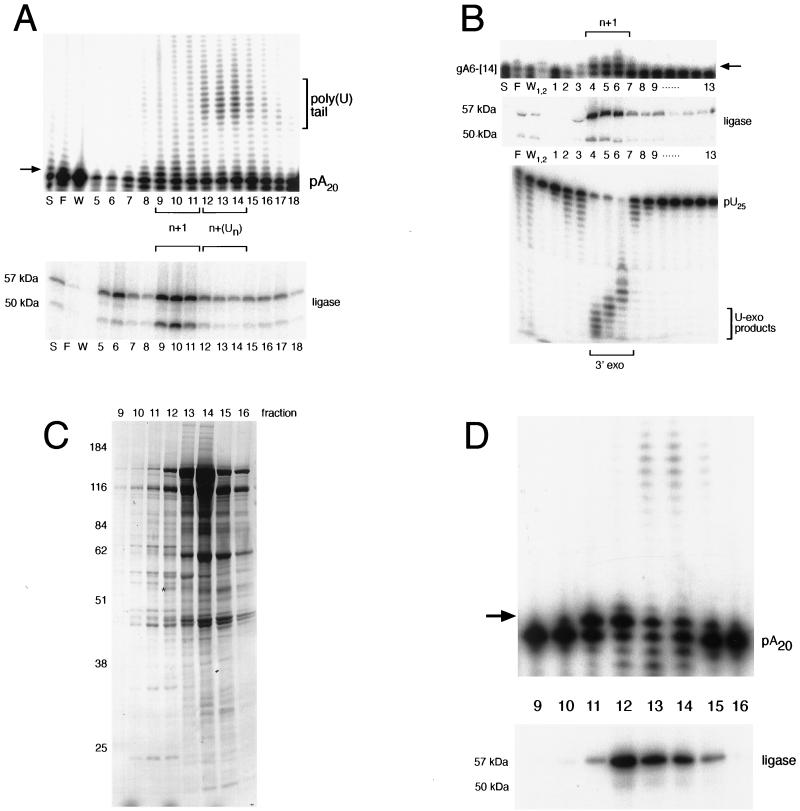
FIG. 4.
Single-U addition activity copurifies with RNA ligase and exonuclease. (A) RNA ligase and multiple-U addition activities are chromatographically distinct on heparin-Sepharose. A 1-pmol sample of 5′-labeled poly(A) RNA (designated pA20) was incubated with 1 μl of each fraction in the presence of 1 mM UTP. RNA ligase in each of the corresponding fractions was adenylated and is shown at the bottom. (B) Single-U addition activity cofractionates with U exonuclease (U-exo) activity. Fractions 9 to 11 from the heparin-Sepharose column (A) were pooled and further purified on Q-Sepharose. A 1-pmol sample of 3′-labeled A6-[14] was incubated with 1 μl of each fraction in the presence of 1 mM UTP. RNA ligase in each of the fractions was adenylated and is shown at the bottom. For the exonuclease assay, 1 pmol of 5′-end-labeled poly(U) RNA (designated pU25) was incubated with 1 μl of each fraction. For the TUTase assays, single-U addition is shown by an arrow. S, F, and W, respectively, indicate starting material, flowthrough, and wash. (C) Silver staining of proteins present in the glycerol gradient purification step. Fractions 4 and 5 from the Q-Sepharose column (B) were pooled, and the complexes were separated on a 10-to-30% glycerol gradient. Following adenylation, protein was isolated and analyzed by SDS–10% PAGE. Size markers are shown at the left, and sizes are given in kilodaltons. Gradient fractions are indicated above. An asterisk indicates the 57-kDa radioactively labeled ligase. (D) Single-U addition products coincide with a stable 19S complex. A 1-pmol sample of 5′-labeled poly(A) RNA was incubated with 1 μl of each glycerol gradient fraction in the presence of 1 mM UTP. RNAs were recovered and analyzed by 8% urea–PAGE. Adenylated ligase that corresponds to these fractions is shown at the bottom.