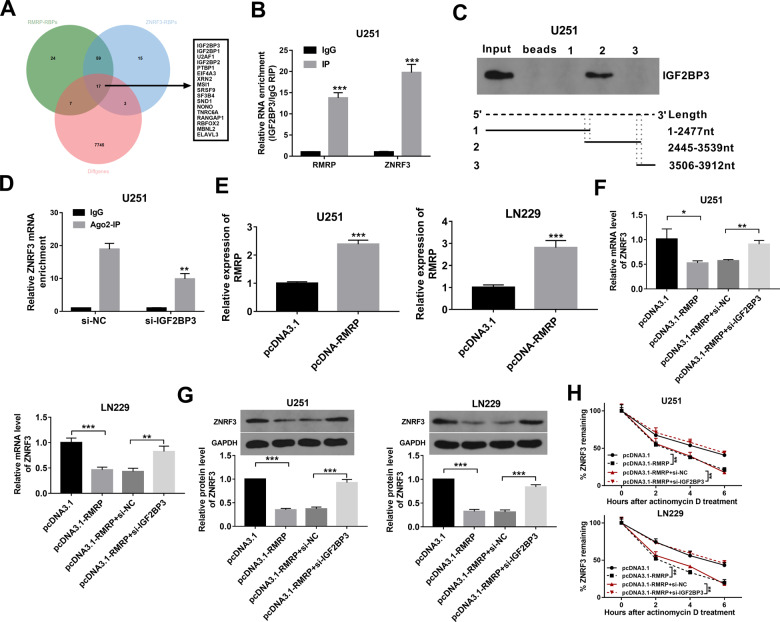
Fig. 6. RMRP regulated ZNRF3 expression and ZNRF3 mRNA stability through RBP IGF2BP3.
A Venn analysis for RMRP-RBPs, ZNRF3-RBPs, and differentially expressed genes in glioma versus normal tissues. RMRP-RBPs (green): RBPs with the possibility to bind with RMRP (predicted by the POSTAR database). ZNRF3-RBPs (blue): RBPs with the possibility to bind with ZNRF3 mRNA (predicted by the POSTAR database). Diffgenes (red): Differentially expressed genes in glioma versus normal brain tissues (data were downloaded from the TCGA/GTEx databases). B RIP assay was performed using IgG or IGF2BP3 antibody in U251 cells. Next, RMRP and ZNRF3 mRNA levels enriched by IgG or IGF2BP3 antibodies were measured by RT-qPCR assay (n = 3). C RNA pull-down assay was performed using biotin-labeled ZNRF3 mRNA 3’UTR fragment 1, fragment 2, and fragment 3 in U251 cells, followed by the detection of IGF2BP3 protein level via western blot assay. D U251 cells were transfected with si-NC or si-IGF2BP3. At 48 h after transfection, a RIP assay was performed using IgG or Ago2 antibody. E U251 and LN229 cells were transfected with pcDNA3.1 or pcDNA3.1-RMRP. Forty-eight hours later, the RMRP level was measured by RT-qPCR assay (n = 3). F, G U251 and LN229 cells were transfected with pcDNA3.1, pcDNA3.1-RMRP, pcDNA3.1-RMRP + si-NC, or pcDNA3.1-RMRP + si-IGF2BP3. F, G ZNRF3 mRNA and protein levels were determined by RT-qPCR and western blot assays (n = 3). H ZNRF3 mRNA stability was tested by Actinomycin D and RT-qPCR assays (n = 3). **P < 0.01. ***P < 0.001.