Abstract
Cells expressing high levels of the cyclin‐dependent kinase (CDK)4/6 inhibitor p16 (p16High) accumulate in aging tissues and promote multiple age‐related pathologies, including neurodegeneration. Here, we show that the number of p16High cells is significantly increased in the central nervous system (CNS) of 2‐year‐old mice. Bulk RNAseq indicated that genes expressed by p16High cells were associated with inflammation and phagocytosis. Single‐cell RNAseq of brain cells indicated p16High cells were primarily microglia, and their accumulation was confirmed in brains of aged humans. Interestingly, we identified two distinct subpopulations of p16High microglia in the mouse brain, with one being age‐associated and one present in young animals. Both p16High clusters significantly differed from previously described disease‐associated microglia and expressed only a partial senescence signature. Taken together, our study provides evidence for the existence of two p16‐expressing microglia populations, one accumulating with age and another already present in youth that could positively and negatively contribute to brain homeostasis, function, and disease.
Keywords: Aging, cellular senescence, senescence, p16, neuroscience
P16+ microglia increases in aging brains of mice and humans. P16+ cells are found in 597 different microglia populations, including two previously unknown clusters.

Abbreviations
- AD
Alzheimer’s disease
- CAM
CNS associated macrophages
- CDK
Cyclin dependent kinase
- CDP
Central animal facility
- CNS
Central nervous system
- DAM
Disease associated microglia
- DEC
Animal care and use committee
- GRP
Glial restricted progenitor
- HOM
Homeostatic microglia
- HSV
Herpes simplex virus
- HVG
Highly variable feature
- IFN
Interferon microglia
- logFC
Log fold change
- mRFP
Monomeric red fluorescent protein
- MS
Multiple sclerosis
- OPC
Oligodendrocyte progenitor cell
- PCA
Principal component analysis
- PD
Parkinson’s disease
- qRT‐PCR
Quantitative real‐time poly chain reaction
- SASP
Senescence associated secretory phenotype
- tTK
thymidine kinase
- UM
Unknown microglia
- UMAP
Uniform manifold approximation and projection
- WGCNA
Weighed gene correlation network analysis
1. INTRODUCTION
Cyclin‐dependent kinase (CDK)4/6 inhibitor p16INK4a (from now on referred to as p16) levels gradually increase with age in multiple tissues and organisms (Herbig et al., 2006; Liu et al., 2009; Melk et al., 2004; Yousefzadeh et al., 2020). p16High cells actively contribute to aging and age‐associated dysfunctions by restricting the regenerative potential of the tissue (Martin et al., 2014) and promoting chronic inflammation (Sanada et al., 2018). Genetic or pharmacological ablation of p16High cells is able to increase health‐ and lifespan in mice (Baker et al., 2016; Xu et al., 2018). p16 expression is a common feature of cellular senescence (Liu et al., 2019), a state of stable and generally irreversible growth arrest originally described as a key process regulating cellular and organismal aging (Hayflick & Moorhead, 1961). Senescent cells are characterized by various structural changes, including misshaped nuclei, enhanced lysosomal content and phagocytic activity, altered mitochondria morphology, and changed plasma membrane composition (Hernandez‐Segura et al., 2018). In addition, senescent cells acquire a pro‐inflammatory phenotype by releasing cytokines and chemokines (a phenotype collectively defined as the SASP—senescence‐associated secretory phenotype) (Gorgoulis et al., 2019). Virtually, all cells can up‐regulate p16 levels, but this induction is not always reflected by a fully senescent state. For example, p16 expression is significantly increased in aged macrophages (Hall et al., 2016), but p16 overexpression can also be observed in young macrophages responding to physiological stimuli (Hall et al.,l., 2017), (Behmoaras & Gil, 2021).
Aging leads to a reduction in brain volume and cognition (Peters, 2006) and is the main risk factor for dementia and neurodegeneration (Wyss‐Coray, 2016). Aging and neurodegenerative conditions induce a common gene expression signature in microglia, the resident immune cells of the CNS (Galatro et al., 2017). Microglia exhibit a hypersensitive and pro‐inflammatory phenotype, known as priming, in particular during aging and neurodegeneration (Norden & Godbout, 2013; Perry & Holmes, 2014; Raj et al., 2014). These primed microglia exert an increased inflammatory response and thereby alter CNS function (Norden & Godbout, 2013). In addition to primed immune cells, the accumulation of pro‐inflammatory senescent cells in the CNS may also predispose elderly to neurodegenerative diseases or aggravate disease etiology (Kritsilis et al., 2018). In the CNS, p16 expression increases during natural aging and in brains affected by pathologies such as Parkinson's disease (PD), multiple sclerosis (MS), and Alzheimer's disease (AD) (Martin‐Ruiz et al., 2020; Nicaiseet al., 2019; Zhang et al., 2019). Removal of p16High cells ameliorates the progression of neurodegeneration in amyloid and tau AD mouse models and in mice exposed to the neurotoxin paraquat (Bussian et al., 2018; Chinta et al., 2018; Zhang et al., 2019). In a neurodegenerative context, different cell types become p16High and influence disease progression. A recent study has attempted to identify senescent cell types naturally occurring in the murine aging brain using single‐cell transcriptomic profiling, and identified an enrichment of p16High cells in microglia and OPCs (Ogrodnik et al.,l., 2021). However, a limitation of single‐cell RNA sequencing (scRNAseq) is its ability to detect low abundant transcripts, which is the case of the p16 transcript. Here, we aimed to identify p16High cell populations in the aging brain by using a transgenic mouse model that allows for the isolation of cells expressing p16 at the protein level, and then perform validation of the findings in wild‐type mice and humans.
2. RESULTS
2.1. RFPHigh cells expressing inflammatory and phagocytosis‐related genes accumulate in the aging brain of p16‐3MR mice
The p16‐3MR mouse contains a monomeric red fluorescent protein (mRFP) fused to Renilla Luciferase and a truncated herpes simplex virus (HSV)‐1 thymidine kinase (tTK), under control of the p16 promoter (Demaria et al., 2014). In order to evaluate whether the levels of the 3MR transgene and the number of 3MRHigh cells increase in the brain with age, we measured RFP signal and percentage of cells expressing high levels of RFP in 7‐ to 12‐week (defined young) and 105‐ to 116‐week (defined old) mice by flow cytometry (Figure S1a). The mean mRFP intensity was significantly higher in old mice (Figure 1a), and the percentage of cells expressing high levels of RFP (RFPHigh) cells increased >sevenfold with aging, from ~0.2% in young to ~1.5% in old mouse brains (Figure 1b). Importantly, the purified RFPHigh population was enriched in cells expressing high levels of the p16 transcript (Figure S1b).
FIGURE 1.
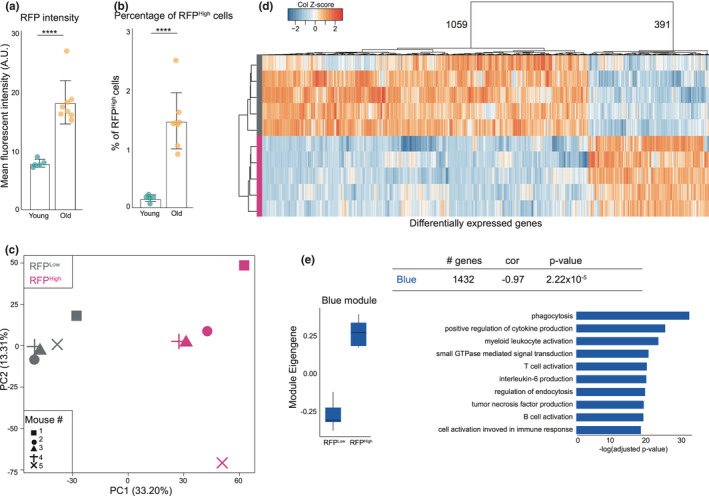
p16‐RFP expression is increased in the brain of aged p16‐3MR mice and abundantly express inflammatory and microglia genes. (a) Mean fluorescent RFP intensity of all viable cells in young compared to old brains. ****p<0,0001. (b) Percentage of viable cells positive for RFP in young mouse brains compared to old. ****p<0,0001. (c) PCA plot of bulk sequenced RFPLow and RFPHigh cells from old mouse brains. (d) Heatmap of all differentially expressed gene between the RFPLow and RFPHigh samples. E: Expression and gene‐ontology analysis of a WGCNA module enriched in RFPHigh samples
We then isolated RFPLow and RFPHigh cells from aged brains and generated gene expression profiles of both populations using bulk RNA sequencing (RNAseq). Principal component analysis (PCA) showed significant transcriptional differences between the RFPLow and RFPHigh populations as indicated by the first principal component (Figure 1c). Differential gene expression analysis revealed 1459 differentially expressed genes between the two populations (Figure 1d). Among the most enriched genes in the RFPHigh samples (Table S1) were Cass4 and Apba2 (or Mint2), which are involved in amyloid synthesis and AD (Beck et al., 2014; Ho et al., 2008) and genes associated with macrophage activation, like Akr1b3, Angptl7, and Ticam2 (Qian et al., 2016; Ramana et al., 2006; Seya et al., 2005).
To determine whether gene networks in RFPHigh samples associated with specific biological or cellular functions, a weighted gene correlation network analysis (WGCNA) (Langfelder & Horvath, 2008) was performed, resulting in branches, or modules, of highly correlating genes (Figure S1c; Table S2). One of these modules (the “blue” module), involved in phagocytosis and cytokine production, was significantly enriched in the RFPHigh samples, as reflected by the Module Eigengene, or first principal component, of the module (Figure 1e; Figure S1d‐h). These data suggest that RFPHigh cells accumulate in the aging brain and are enriched in expression of genes associated with inflammation and phagocytosis pathways.
2.2. Single‐cell transcriptomic profiling demonstrates accumulation of RFPHigh microglia with aging in p16‐3MR mice
To further characterize the phenotype of the RFPHigh cell population in the aged mouse CNS, we compared scRNAseq profiles of purified RFPHigh cells to unsorted CNS cell samples (Figure S2a‐d; Table S3). We identified 14 clusters in the dataset, using unsupervised, graph‐based clustering analysis where each cluster corresponds to a distinct cell type (Figure 2a). The cell types were identified based on the expression of well‐known cell type marker genes: P2ry12, Cx3cr1, and Tgfbr1 for microglia; Cldn5 for endothelial cells; Gfap, Aqp4, and Atp1b2 for astrocytes; Grid2 for Purkinje neurons; Npy and Fabp7 for glial restricted progenitors (GRP); Cd3g for T/NK cells; H2‐Aa for monocytes; F13a1 for CNS‐associated macrophages (CAMs); Pdgfrb for mural cells; Acta2 for neutrophils; Map1b for neurons; Dcn and Col1a1 for fibroblasts; Olig1, Mobp, and Plp1 for oligodendrocytes; Ms4a1 for B cells; Ttr for unidentified population 1 (unknown 1); and Ak7 for unidentified population 2 (unknown 2) (Figure 2b; Table S4). Next, for the total viable and the RFPHigh populations, the distribution of cell types within each sample was compared. Microglia, astrocytes, and endothelial cells were the most abundant cell types obtained with our isolation method (total viable population) from aged mouse brains, while other cell types such as neurons and oligodendrocytes were less abundant, and most likely underrepresented compared to their normal physiological distribution in the CNS (Valério‐Gomes et al., 2018). Strikingly, the RFPHigh sample was almost exclusively comprised of microglia (94.6%) and some glial restricted progenitors (2.6%) (Figure 2c and d).
FIGURE 2.
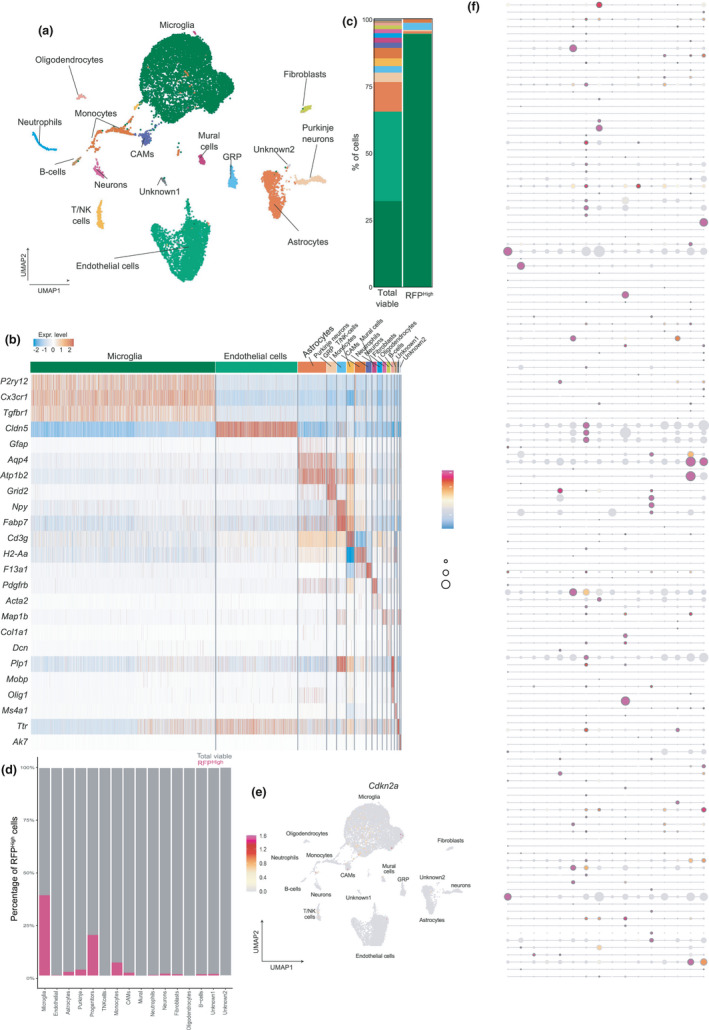
RFPHigh cells are highly enriched for microglia. (a) UMAP depicting mouse CNS with cluster annotations based on cell types. (b) Heatmap showing the expression of cell type markers in each cluster. (c) Barplot of cluster distribution of total viable cells and RFPHigh cells. (d) Barplot showing the percentage of RFPHigh cells for each cell type. (e) Cdkn2a plotted in UMAP of all sequenced single cells. (f) Dotplot showing the expression of senescence markers in each cluster
The scRNAseq data confirmed that microglia expressed Cdkn2a, the genomic locus containing p16, more abundantly compared to other cell types in the CNS (Figure 2e). To investigate whether microglia showed additional markers of cellular senescence, the expression levels of a list of 162 senescence‐associated genes in each cell type were evaluated (Table S5). These genes were variably expressed and not abundantly present in the microglia population (Figure 2f). These data suggest that RFPHigh microglia accumulate in the aging brain of p16‐3MR mice and that their transcriptional profile differs from a classical senescence‐associated gene signature.
2.3. Microglia are enriched in p16 in the brains of wild‐type mice and humans
To confirm the presence of RFPHigh microglia in aged brains, we used different methods. First, from the bulk RNAseq list, we investigated the expression level of cell type‐specific genes in the RFPHigh fraction: Hexb, Cxcr1, P2ry12, and Tmem119 for microglia; Aqp4 and Gfap for astrocytes; Cldn5 and Vcan for endothelial cells; Rbfox3 for neurons; F13a1 for CNS‐associated macrophages; Plp1 for oligodendrocytes; and Pdgfra for oligodendrocyte progenitor cells and fibroblasts (Figure 3a). The expression level of microglia genes was consistently higher in the RFPHigh samples, while in the RFPLow samples, endothelial cell, oligodendrocyte, and oligodendrocyte progenitor cell markers were more abundantly expressed. Second, we deconvoluted transcriptomes of the bulk RFPHigh samples with CIBERSORT, using our single‐cell data as the reference matrix (Table S4). Again, a pattern of enrichment for microglia in the RFPHigh cell population was observed (Figure 3b).
FIGURE 3.

Increased expression of p16 in mouse and human microglia. (a) Gene expression of cell marker genes in RFPLow compared to RFPHigh mouse samples. (b) Barplot showing the distribution of cells types in the mouse CNS bulk dataset after deconvolution. (c) p16 expression measured by qPCR in cells isolated from young and old mouse brains. ****p<0,0001. (d) CCKN2A expression in human microglia and total cortical tissue (from Galatro et al., 2017). ****p<0,0001E: UMAP depicting CDKN2A expression in 450,000 CNS cell nuclei (Gerrits et al. 2021)
To validate the correlation between p16 and RFP positivity in a non‐transgenic background, we measured p16 levels in wild‐type animals. We isolated microglia, astrocytes, and non‐microglia/non‐astrocyte (defined as “the rest”) cells from the brain of young and old wild‐type C57BL/6 mouse brains and evaluated the p16 transcript levels of the isolated populations. Only microglia of old mice revealed a significant p16 upregulation, while no significant differences between young and old mice were detected neither in astrocytes, a cell population that was minimally represented in the RFPHigh cells isolated from aged p16‐3MR mice, nor in other mixed cell types mainly consisting of endothelial cells (Figure 3c).
Next, we evaluated the level of p16 expression in human microglia and cortical CNS tissue (Galatro et al., 2017). Strikingly, we measured a significant enrichment for CDKN2A, the genomic locus containing p16, in the microglia population compared to the total brain samples (Figure 3d). In addition, we determined the expression levels of CDKN2A in a single‐nucleus RNA sequencing data set of human AD cases and healthy donors (Gerrits et al., 2021). Also in this dataset, CDKN2A was most abundantly expressed by microglia (Figure 3e). Interestingly, lymphocytes and oligodendrocytes, underrepresented in our mouse scRNAseq, also expressed CDKN2A in human brains. Altogether, these data confirm that both in the mouse and in the human aged brain, p16High cells are mostly present in the microglia population.
2.4. RFPHigh cells cluster in two distinct and previously unreported microglia populations
Recent reports based on single‐cell transcriptomes identified context‐dependent microglia subtypes (Masuda et al., 2020; Sierksma et al., 2020). Subclustering analysis of the entire microglia population from our single‐cell dataset (RFPHigh and unpurified) revealed 5 distinct subpopulations: 3 previously described—a population which surveils the surroundings and maintains homeostasis through clearance of cellular debris, called homeostatic (HOM); a more reactive population, which acquires pro‐inflammatory and antigen‐presenting properties, called disease‐associated microglia (DAM); and activated microglia with high interferon signaling (IFN)—and 2 additional clusters, named unknown microglia clusters 1 and 2 (UM1 and UM2), which segregated from the known clusters and were almost exclusively derived from the RFPHigh samples (Figure 4a; Figure S3a). The HOM cluster was depleted in the RFPHigh microglia, while DAM and IFN clusters were equally present in both RFPHigh and RFPLow populations. Differential gene expression analysis revealed a clear distinction of the RFPHigh microglia from the total viable population (Figure 4b), even if the expression of selected senescence‐associated genes was not specifically enriched in the UM1 and UM2 clusters, but seems to be slightly increased in the DAM cluster (Figure 4c; Figure S3d). Single‐cell regulatory network inference and clustering (SCENIC) analysis identified 43 gene networks differentially expressed between RFPHigh and total microglia. Interestingly, expression of genes regulated by Ets2, a transcription factor that positively regulates p16 expression (Kotake et al., 2015), was enriched in RFPHigh microglia (Figure 4d; Figure S3b).
FIGURE 4.

p16High microglia express genes associated with inflammation, cell cycle response, and cell motility. (a) UMAP plots where colors indicate the different clusters within all the sequenced microglia cells. DAM=damage‐associated microglia. (b) Volcano plot depicting differential expressed genes between the RFPHigh microglia and total viable microglia. (c) Violin plot showing the expression of senescence genes in each microglia cluster. (d) Heatmap showing the differentially expressed regulons in the SCENIC analysis between all RFPHigh and total viable microglia. (e) GOs significantly enriched in the p16‐UM1 cluster. (f) GOs significantly enriched in the p16‐UM2 cluster. (g) Violin plot depicting the expression of UM1 and UM2 cluster markers with age in wild‐type mice of the dataset from Zhang et al. (Zhang et al., 2020)
We then investigated the predicted functions of genes upregulated in the RFPHigh microglia. In line with our bulk RNAseq results, two AD risk genes were upregulated in the RFPHigh microglia. Gsap selectively increases amyloid‐beta production (He et al., 2010), a protein that is aggregated in AD and inositol polyphosphate‐5‐phosphatase D (Inpp5d) is suggested to contribute to AD in a non‐amyloid‐beta‐dependent fashion (Efthymiou & Goate, 2017). Additionally, we found genes involved in macrophage motility and myelination. Plxnb2 has been shown to negatively regulate cell motility (Roney et al., 2011), while Kif13b regulates myelination in the CNS (Noseda et al., 2016) (Table S4). In addition, we examined the genes upregulated in each UM cluster. Gene ontology analysis for genes enriched in the UM1 cluster showed an enrichment for genes involved in the ERK/MAPK pathways (Figure 4e) suggested to underlie CNS inflammation (Kaminska et al., 2009). Genes highly expressed in UM2 microglia were associated with cell cycle response and Rho GTPase signaling (Figure 4f), a pathway necessary for process motility, which is important for scanning of the parenchyma (Neubrand et al., 2014).
Finally, we compared the gene expression profile of the RFPHigh microglia to previously reported disease‐ and aging‐associated microglia profiles (Table S5). While both the DAM and the IFN clusters significantly overlap with previously reported profiles, none of the investigated gene sets was significantly enriched in our UM1 and UM2 clusters (Figure S3c). Interestingly, when we looked at the expression levels of UM1 and UM2 cluster marker genes in aging wild‐type mice from the dataset of Zhang et al. 2020, we observed that UM1 cluster markers were expressed in microglia at all ages albeit lower at 19 months, while the expression of UM2 cluster marker genes progressively increased with age in these wild‐type mice (Figure 4g). In summary, these data show that RFPHigh microglia cluster in two distinct subpopulations with previously unreported gene signatures which we named UM1 and UM2. UM1 negatively correlates with age and is characterized by expression of inflammatory genes. In contrast, UM2 is age‐associated and characterized by differential expression of genes involved in cell cycle regulation and cell motility.
3. DISCUSSION
Microglia, tissue‐resident macrophages of the CNS, is a heterogeneous cell population that change over the course of an organism lifespan. Microglia heterogeneity decreases with age, but several states—for example chemokine‐enriched inflammatory microglia—remain unchanged or increase in aged brains (Hammond et al., 2019). Moreover, microglia are reported to age in a regional‐dependent manner (Grabert et al., 2016). However, there is still little understanding of the phenotypical characteristics of microglia subpopulations in the aged brain. The current study reveals two previously unreported p16‐expressing microglia subpopulations, one with a quite stable expression across different life stages and one which accumulation significantly increases with age.
Elevated p16 expression is a marker of cellular senescence and has been used to identify the accumulation of senescent astrocytes (Bhat et al., 2012; Chinta et al., 2018; Yabluchanskiy et al., 2020), oligodendrocyte progenitor cells (Nicaise et al., 2019; Zhang et al., 2019), and neurons in the human aging brain (Kang et al., 2015) and in mouse models of neurodegeneration. Moreover, recent data indicated that microglia accumulate p16High cells in aged mouse brains (Ogrodnik et al., 2021).
In this study, using both transgenic and wild‐type mice, and various publicly available mouse and human transcriptomic datasets, we identified two distinct subpopulations of p16High microglia, one constantly present and one age‐associated, that did not express a classical senescence‐associated gen signature. Absence of a senescence profiling is in line with a previous study showing that while murine microglia in vitro show markers of replicative senescence, the microglia of aged mice express higher levels of p16 but not other typical senescence‐associated changes (Stojiljkovic et al., 2019).
Distinct transcriptional changes in each cell population were found during single‐cell sequencing of the aged murine brain (Ximerakis et al., 2019), indicating that each cell type ages differently. In our single‐cell study, only astrocytes, endothelial cells, and microglia were represented in large quantities, while other cell types were underrepresented due to our cold protease isolation procedure. Since we also identified higher expression of CDKN2A in lymphocytes and oligodendrocytes by analyzing a dataset derived from RNAseq of single nuclei isolated from human brains (Gerrits et al., 2021), it remains to be seen whether other less represented populations also express p16 with age.
Our data suggest a clear separation of the p16High microglia from other microglia populations and the existence of two distinct subsets—one expressed across the entire lifespan and the other age‐associated. A subset of p16High microglia may be part of a homeostatic mechanism aimed at reducing damage propagation, via cell cycle arrest and improved phagocytic properties, and at promoting immune surveillance, via activation of specific secretory and pro‐inflammatory phenotypes. On the other side, the accumulation of a subset of p16High cells with age may represent the byproduct of excessive damage and reduced clearance capacity, which could contribute to detriment accumulation and loss of tissue homeostasis. Future studies need to address this issue by evaluating the effects of specifically eliminating specific p16High microglia subsets, and to further characterize the presence and function of these subsets in the human brain. It will also be important to evaluate whether current senolytic approaches are eliminating these p16High microglia subsets, and the balance between benefits and toxicities of removing such populations.
4. MATERIALS AND METHODS
4.1. Mice
p16–3MR mice with a C57BL/6 background or wild‐type C57BL/6 were used for all experiments (Demaria et al., 2014). Young mice were between 7 and 12 weeks of age, and old mice were between 105 and 116 weeks of age. The young mice were a mix of males and females (n=5), male old mice were used for bulk sequencing (n=5), and female mice were used for single‐cell sequencing (n=4). Young, 18 weeks of age, (n=3) and old, 101 and 104 weeks of age, (n=3) wild‐type mice were used for the isolation of astrocytes, microglia, and rest cells. Mice were raised on a 12‐hr light/dark cycle with food and water available ad libitum and were individually housed. All experiments were performed in the Central Animal Facility (CDP) of the UMCG, with protocol (15339–02–001) approved by the Animal Care and Use Committee (DEC) of the University of Groningen.
4.2. Cell isolation from mouse brain tissue
Cells were isolated from adult mouse brain using an enzymatic protocol at 4℃. The brains were isolated and dissociated by three rounds of GentleMACS (m_brain_01, m_brain_02, and m_brain_03) in enzyme mix of 15 mg/ml Protease (Sigma P5380), 1 mM L‐cysteine hydrochloride (Sigma C7477), and 0.5 µg/µl DNase (Roche 10104159001) with 10 min incubation in the mix on ice in between GentleMACS programs. The homogenized brain samples were passed through a 100 μM cell strainer to obtain a single‐cell suspension. The cells were centrifuged at 300 rcf for 10 min at 4℃, and the pellet was resuspended in 24% Percoll gradient buffer. 3 mL dPBS was pipetted onto the gradient buffer, and myelin was removed by centrifuging at 950 rcf for 20 min at 4℃. The cell pellets were incubated with DAPI and Draq5. Viable cells were FACS sorted as DAPInegDraq5pos events. RFPHigh and RFPLow bulk samples were sorted from individual mice, but for the single‐cell sequencing, RFPHigh (21,500) and total viable cells (45,000) from four mice were combined each into one lane of a 10X Genomics Chromium chip.
For the isolation of astrocytes, microglia, and rest cells, cell pellets were incubated with the antibodies CD11b‐BV421 (clone M1/70, Biolegend, San Diego, CA, USA), CD45‐FITC (clone 30‐F11, Biolegend, San Diego, CA, USA), CD49d‐PE (clone R1‐2, Miltenyi Biotec), Acsa2‐FITC (clone REA969, Miltenyi Biotec), PI, and Draq5. Microglia were FACS sorted as PIneg Draq5pos CD11bhigh CD45int CD49dneg events. Astrocytes were FACS sorted as PIneg Draq5pos CD11bneg CD45neg Acsa2pos events and rest cells as PIneg Draq5pos CD11bneg CD45neg Acsa2neg events. Bulk samples were sorted from individual mice.
4.3. FACS analysis
Flowjo V.10 was used to analyze the mean, median RFP expression, number of RFP positive cells, and viability of cells. Unpaired t tests were used to compare the mean, median, and number of positive cells. Paired t test was used to compare viability.
4.4. Real‐Time PCR
Total RNA was prepared using the AllPrep DNA/RNA Micro Kit (Qiagen, 80284). RNA was reverse transcribed into cDNA using a kit (Applied Biosystems). Quantitative RT‐PCR (qRT‐PCR) reactions were performed as described (Demaria et al., 2010) using the Universal Probe Library system (Roche). Primer used:
mp16 #91 ‐FAATCTCCGCGAGGAAAGC ‐RGTCTGCAGCGGACTCCAT.
mHprt1 #62 ‐FATCACATTGTGGCCCTCTG ‐RGTCATGGGAATGGATCTATCACT.
mHmbs #91 ‐FAGAAAAGTGCCGTGGGAAC ‐RTGTTGAGGTTTCCCCGAAT.
4.5. Bulk RNAseq library construction and sequencing
RNA was isolated from cell pellets with the AllPrep DNA/RNA Micro Kit (Qiagen, 80284). RNA concentrations were measured on a Qubit using a HS RNA kit. 2,5 ng of the samples was used for library preparation with the Lexogen QuantSeq 3’ mRNA‐Seq Library Prep Kit (FWD) from Illumina. All libraries were pooled equimolarly and sequenced on a NextSeq 500 at the sequencing facility in the UMCG.
4.6. scRNAseq library construction and sequencing
The single‐cell cDNA libraries were constructed using the Chromium Single Cell 3’ Reagents Kit v3 and corresponding user guide (10x Genomics). All samples were pooled in equimolar ratios and sequenced on a NextSeq 500 at the sequencing facility in the UMCG.
4.7. Gene sets from literature
To compare our microglia clusters with reported microglia phenotypes in literature, several gene sets were downloaded. From (Sierksma et al., 2020), EV7 was downloaded and genes with a p_val_adj <0.05 and logFC >0.15 were selected (304 genes) and from EV6 the CPM gene set (521 genes). From (Hammond et al., 2019), table S1 was downloaded and marker genes from clusters OA2 and OA3 were selected (136 and 37 genes, respectively). From (Keren‐Shaul et al., 2017), table S2 was downloaded and upregulated genes of “Microglia3” with a p_val_adj <0.05 were selected (469 genes). From (Butovsky & Weiner, 2018), upregulated genes listed in Figure 2 were used (29 genes). From (Gerrits et al., 2020), genes from table S4 with a p_val_adj <0.05 and logFC >0.15 were selected (188 genes). From Galatro et al. (2017), Voom Normalized counts were downloaded from GEO. From Gerrits et al. 2021, the exact same analyzed data objects as reported in the paper were used as these were generated by ourselves.
4.8. Bulk RNAseq data analysis
Data preprocessing was performed with the Lexogen Quantseq 2.3.1 FWD UMI pipeline on the BlueBee Genomics Platform (1.10.18). Count files were loaded into R, and DAFS filtering was performed to remove lowly expressed genes (George & Chang, 2014). A negative binomial generalized log‐linear model was used to model gene expression levels, as implemented in edgeR, adjusted for mouse since the RFPLow and RFPHigh cells were obtained from the same mice and differentially expressed genes were determined using a likelihood ratio test (Robinson et al., 2010). Thresholds were set at abs(logFC) >1 and p < 0.05. Principal component analysis was performed on logCPM transformed counts. Visualizations were made with the CRAN package “ggplot2.” Heatmaps were made with the CRAN package “gplots,” and rows and columns were clustered using hierarchical clustering with the ward.D2 method on Pearson's correlations. For WGCNA analysis, VST‐transformed counts obtained from DESeq2 were used as input (Langfelder & Horvath, 2008; Love et al., 2014). Signed WGCNA was performed using biweight mid‐correlations, and the max number of excluded outliers was restricted to 10%. Since we were dealing with binary data (i.e., two experimental groups), the robust treatment for the y variable of the biweight mid‐correlation was turned off (Langfelder & Horvath, 2012). Gene ontology analysis was performed on significantly differentially expressed genes (p < 0.05 and logFC >0.15) using “clusterProfiler” with a p‐ and q‐value cutoff of 0.05.
4.9. scRNAseq data analysis
Raw reads were processed using Cell Ranger 3.0.0 with default settings and aligned to the mouse mm10 genome. Barcode filtering was performed with DropletUtils with a threshold on >250 UMIs. Counts from cellular barcodes were then extracted from the raw output count matrix from Cell ranger. Cells with a mitochondrial content >10% were removed from the dataset. Counts from the different sample groups were merged into one using the “Merge” function from Seurat (v3). Then, the data were SCTransformed with regression on mitochondrial and ribosomal content, and subsequently, PCA, UMAP, finding neighbors, and clustering were performed as implemented by Seurat (Hafemeister & Satija, 2019). For differential gene expression analysis, raw counts were normalized using the “NormalizeData” function; then, DE genes were identified with MAST. Geneset scores were calculated using the “AddModuleScore” function. Average gene expression per cluster was calculated using the “AverageExpression” function. Median of expressed genes that were mitochondrial per cell: 2.2%; ribosomal: 5.6%; and median number of genes detected per cell: 755.
Regulatory gene network (regulon) analysis was performed using SCENIC; normalized counts from Seurat were used as input (Aibar et al., 2017). Only genes with more than 3 counts and present in at least 0.5% of all cells were included. GENIE3 and SCENIC were used with default settings (Huynh‐Thu et al., 2010; Aibar et al., 2017). Enrichment of gene sets and regulons in our scRNAseq data was quantified using AUCell. AUC values are plotted as an average per group. Regulons with a median AUC <0.01 were excluded in the downstream analysis.
From Zhang et al. (2020), the raw count matrices of all mice were downloaded and raw reads were processed using Cell Ranger 3.0.0 with default settings and the pre‐mRNA package. From the bam file, exonic reads and intronic reads mapping in the same direction as the mRNA were counted per barcode with Abacus in order to distinguish barcodes containing nuclear RNA from ambient and cytoplasmic RNA (Xi et al., 2020). The counts corresponding to these barcodes were extracted from the raw count matrix generated by Cell Ranger and loaded in R with Seurat (3.0.3). Nuclei with a mitochondrial content >5% were removed from the dataset. Count matrices of all mice were merged. The data were normalized for library size, by a scale factor of 10,000 and log‐transformed. Scrublet was used to identify and remove doublets (Wolock et al., 2019) (Wolock et al., 2019). Highly variable features (HVGs) were determined using the VST method. The data were scaled and heterogeneity associated with number of UMIs and mitochondrial content was regressed out and the data were clustered using the graph‐based clustering approach implemented in Seurat. The microglia cluster was identified based on expression of P2ry12, Csf1r, and Cx3cr1. Then, only WT mice were used for further analysis. Geneset scores were calculated using the “AddModuleScore” function from Seurat.
CONFLICT OF INTEREST
MD is co‐founder, shareholder, and advisor for Cleara Biotech. The project was not funded or influenced by Cleara.
AUTHOR CONTRIBUTIONS
N.T., E.G., and B.W. involved in methodology. N.T. and E.G. involved in validation and formal analysis. N.T., E.G., B.W., B.E., and M.D. involved in investigation. N.T., B.E., and M.D. involved in writing–original draft preparation, conceptualization, and writing–review and editing. B.E. and M.D. involved in supervision and funding acquisition.
Supporting information
Figure S1‐S3
Table S1‐S7
ACKNOWLEDGMENTS
We thank the Demaria and Eggen laboratories for fruitful discussion and Michela Borghesan for technical assistance.
Talma, N. , Gerrits, E. , Wang, B. , Eggen, B. J. L. , & Demaria, M. (2021). Identification of distinct and age‐dependent p16High microglia subtypes. Aging Cell, 20, e13450. 10.1111/acel.13450
Funding information
Funding was provided by the University Medical Center Groningen (UMCG)
Contributor Information
Bart J.L. Eggen, Email: b.j.l.eggen@umcg.nl.
Marco Demaria, Email: m.demaria@umcg.nl.
Data Availability Statement
RNAseq data are deposited in the database GEO (www.ncbi.nih.gov/geo/) with identifier GSE151459. All the data presented here are available from the corresponding authors upon reasonable request.
REFERENCES
- Aibar, S. , González‐Blas, C. B. , Moerman, T. , Huynh‐Thu, V. A. , Imrichova, H. , Hulselmans, G. , Rambow, F. , Marine, J.‐C. , Geurts, P. , Aerts, J. , van den Oord, J. , Atak, Z. K. , Wouters, J. , & Aerts, S. (2017). SCENIC: Single‐cell regulatory network inference and clustering. Nature Methods, 14(11), 1083. 10.1038/NMETH.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D. J. , Childs, B. G. , Durik, M. , Wijers, M. E. , Sieben, C. J. , Zhong, J. , A. Saltness, R. , Jeganathan, K. B. , Verzosa, G. C. , Pezeshki, A. , Khazaie, K. , Miller, J. D. , & van Deursen, J. M. (2016). Naturally occurring p16 Ink4a‐positive cells shorten healthy lifespan. Nature, 530(7589), 184–189. 10.1038/nature16932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T. N. , Nicolas, E. , Kopp, M. C. , & Golemis, E. A. (2014). Adaptors for disorders of the brain? The cancer signaling proteins NEDD9, CASS4, and PTK2B in Alzheimer’s disease. Oncoscience, 1(7), 486–503. 10.18632/oncoscience.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behmoaras, J. , & Gil, J. (2021). Similarities and interplay between senescent cells and macrophages. Journal of Cell Biology, 220(2), 10.1083/jcb.202010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, R. , Crowe, E. P. , Bitto, A. , Moh, M. , Katsetos, C. D. , Garcia, F. U. , Johnson, F. B. , Trojanowski, J. Q. , Sell, C. , & Torres, C. (2012). Astrocyte Senescence as a Component of Alzheimer’s Disease. PLoS One, 7(9), e45069. 10.1371/journal.pone.0045069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussian, T. J. , Aziz, A. , Meyer, C. F. , Swenson, B. L. , van Deursen, J. M. , & Baker, D. J. (2018). Clearance of senescent glial cells prevents tau‐dependent pathology and cognitive decline. Nature, 1. 10.1038/s41586-018-0543-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky, O. , & Weiner, H. L. (2018). Microglial signatures and their role in health and disease. Nature Reviews Neuroscience, 19(10), 622–635. 10.1038/s41583-018-0057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinta, S. J. , Woods, G. , Demaria, M. , Rane, A. , Zou, Y. , McQuade, A. , Rajagopalan, S. , Limbad, C. , Madden, D. T. , Campisi, J. , & Andersen, J. K. (2018). Cellular Senescence Is Induced by the Environmental Neurotoxin Paraquat and Contributes to Neuropathology Linked to Parkinson’s Disease. Cell Reports, 22(4), 930–940. 10.1016/j.celrep.2017.12.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria, M. , Giorgi, C. , Lebiedzinska, M. , Esposito, G. , D'Angeli, L. , Bartoli, A. , Gough, D. J. , Turkson, J. , Levy, D. E. , Watson, C. J. , Wieckowski, M. R. , Provero, P. , Pinton, P. , & Poli, V. (2010). A STAT3‐mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging, 2(11), 823–842. 10.18632/aging.100232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria, M. , Ohtani, N. , Youssef, S. A. , Rodier, F. , Toussaint, W. , Mitchell, J. R. , Laberge, R.‐M. , Vijg, J. , Van Steeg, H. , Dollé, M. E. T. , Hoeijmakers, J. H. J. , de Bruin, A. , Hara, E. , & Campisi, J. (2014). An Essential Role for Senescent Cells in Optimal Wound Healing through Secretion of PDGF‐AA. Developmental Cell, 31(6), 722–733. 10.1016/j.devcel.2014.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiou, A. G. , & Goate, A. M. (2017). Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Molecular Neurodegeneration, 43. 10.1186/s13024-017-0184-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro, T. F. , Holtman, I. R. , Lerario, A. M. , Vainchtein, I. D. , Brouwer, N. , Sola, P. R. , Veras, M. M. , Pereira, T. F. , Leite, R. E. P. , Möller, T. , Wes, P. D. , Sogayar, M. C. , Laman, J. D. , den Dunnen, W. , Pasqualucci, C. A. , Oba‐Shinjo, S. M. , Boddeke, E. W. G. M. , Marie, S. K. N. , & Eggen, B. J. L. (2017). Transcriptomic analysis of purified human cortical microglia reveals age‐associated changes. Nature Neuroscience, 20(8), 1162–1171. 10.1038/nn.4597 [DOI] [PubMed] [Google Scholar]
- George, N. I. , & Chang, C.‐W. (2014). DAFS: a data‐adaptive flag method for RNA‐sequencing data to differentiate genes with low and high expression. BMC Bioinformatics, 15(1), 92. 10.1186/1471-2105-15-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits, E. , Brouwer, N. , Kooistra, S. M. , Woodbury, M. E. , Vermeiren, Y. , Lambourne, M. , Mulder, J. , Kummer, M. , Möller, T. , Biber, K. , Dunnen, W. F. A. D. , De Deyn, P. P. , Eggen, B. J. L. , & Boddeke, E. W. G. M. (2021). ‘Distinct amyloid‐β and tau‐associated microglia profiles in Alzheimer’s disease’, Acta Neuropathologica . Springer Science and Business Media Deutschland GmbH, 141(5), 681–696. 10.1007/s00401-021-02263-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits, E. , Heng, Y. , Boddeke, E. W. G. M. , & Eggen, B. J. L. (2020). Transcriptional profiling of microglia; current state of the art and future perspectives. Glia, 68(4), 740–755. 10.1002/glia.23767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis, V. , Adams, P. D. , Alimonti, A. , Bennett, D. C. , Bischof, O. , Bishop, C. , Campisi, J. , Collado, M. , Evangelou, K. , Ferbeyre, G. , Gil, J. , Hara, E. , Krizhanovsky, V. , Jurk, D. , Maier, A. B. , Narita, M. , Niedernhofer, L. , Passos, J. F. , Robbins, P. D. , … Demaria, M. (2019). Cellular Senescence: Defining a Path Forward. Cell, 813–827. 10.1016/j.cell.2019.10.005 [DOI] [PubMed] [Google Scholar]
- Grabert, K. , Michoel, T. , Karavolos, M. H. , Clohisey, S. , Baillie, J. K. , Stevens, M. P. , Freeman, T. C. , Summers, K. M. , & McColl, B. W. et al (2016). Microglial brain regionâ ’dependent diversity and selective regional sensitivities to aging. Nature Neuroscience, 19(3), 504–516. 10.1038/nn.4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister, C. , & Satija, R. (2019). Normalization and variance stabilization of single‐cell RNA‐seq data using regularized negative binomial regression. Genome Biology. Biomed Central, 20(1), 296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, B. M. , Balan, V. , Gleiberman, A. S. , Strom, E. , Krasnov, P. , Virtuoso, L. P. , Rydkina, E. , Vujcic, S. , Balan, K. , Gitlin, I. I. , Leonova, K. I. , Consiglio, C. R. , Gollnick, S. O. , Chernova, O. B. , & Gudkov, A. V. (2017). p16(Ink4a) and senescence‐associated β‐galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging, 9(8), 1867–1884. 10.18632/aging.101268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, B. M. , Balan, V. , Gleiberman, A. S. , Strom, E. , Krasnov, P. , Virtuoso, L. P. , Rydkina, E. , Vujcic, S. , Balan, K. , Gitlin, I. , Leonova, K. , Polinsky, A. , Chernova, O. B. , & Gudkov, A. V. (2016). Aging of mice is associated with p16(Ink4a)‐ and β‐galactosidasepositive macrophage accumulation that can be induced in young mice by senescent cells. Aging, 8(7), 1294–1315. 10.18632/aging.100991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, T. R. , Dufort, C. , Dissing‐Olesen, L. , Giera, S. , Young, A. , Wysoker, A. , Walker, A. J. , Gergits, F. , Segel, M. , Nemesh, J. , Marsh, S. E. , Saunders, A. , Macosko, E. , Ginhoux, F. , Chen, J. , Franklin, R. J. M. , Piao, X. , McCarroll, S. A. , & Stevens, B. (2019). Single‐Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell‐State Changes. Immunity, 50(1), 253–271.e6. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick, L. , & Moorhead, P. S. (1961). The serial cultivation of human diploid cell strains. Experimental Cell Research, 25(3), 585–621. 10.1016/0014-4827(61)90192-6 [DOI] [PubMed] [Google Scholar]
- He, G. , Luo, W. , Li, P. , Remmers, C. , Netzer, W. J. , Hendrick, J. , Bettayeb, K. , Flajolet, M. , Gorelick, F. , Wennogle, L. P. , & Greengard, P. (2010). Gamma‐secretase activating protein is a therapeutic target for Alzheimer’s disease. Nature, 467(7311), 95–98. 10.1038/nature09325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig, U. et al (2006). Cellular senescence in aging primates. Science, 311(5765), 1257. 10.1126/science.1122446 [DOI] [PubMed] [Google Scholar]
- Hernandez‐Segura, A. , Nehme, J. , & Demaria, M. (2018). Hallmarks of Cellular Senescence. Trends in Cell Biology, 28(6), 436–453. 10.1016/J.TCB.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Ho, A. , Liu, X. , & Südhof, T. C. (2008). Deletion of Mint proteins decreases amyloid production in transgenic mouse models of Alzheimer’s disease. Journal of Neuroscience, 28(53), 14392–14400. 10.1523/JNEUROSCI.2481-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh‐Thu, V. A. , Irrthum, A. , Wehenkel, L. , & Geurts, P. (2010). ‘Inferring Regulatory Networks from Expression Data Using Tree‐Based Methods. PLoS One, 5(9), e12776. 10.1371/journal.pone.0012776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska, B. et al (2009). MAPK Signal Transduction Underlying Brain Inflammation and Gliosis as Therapeutic Target. The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology, 292(12), 1902–1913. 10.1002/ar.21047 [DOI] [PubMed] [Google Scholar]
- Kang, C. , Xu, Q. , Martin, T. D. , Li, M. Z. , Demaria, M. , Aron, L. , Lu, T. , Yankner, B. A. , Campisi, J. , & Elledge, S. J. (2015). ‘The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4’, Science. American Association for the. Advancement of Science, 349(6255), aaa5612. 10.1126/science.aaa5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren‐Shaul, H. , Spinrad, A. , Weiner, A. , Matcovitch‐Natan, O. , Dvir‐Szternfeld, R. , Ulland, T. K. , David, E. , Baruch, K. , Lara‐Astaiso, D. , Toth, B. , Itzkovitz, S. , Colonna, M. , Schwartz, M. , & Amit, I. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell, 169(7), 1276–1290.e17. 10.1016/J.CELL.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kotake, Y. et al (2015). Transcriptional regulation of the p16 tumor suppressor gene. Anticancer Research, 4397–4402. [PubMed] [Google Scholar]
- Kritsilis, M. , V. Rizou, S. , Koutsoudaki, P. , Evangelou, K. , Gorgoulis, V. , & Papadopoulos, D. (2018). Ageing, Cellular Senescence and Neurodegenerative Disease. International Journal of Molecular Sciences, 19(10), 2937. 10.3390/ijms19102937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9(1), 559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2012). Fast R Functions for Robust Correlations and Hierarchical Clustering. Journal of Statistical Software, 46(11), 1–17. 10.18637/jss.v046.i11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. Y. , Souroullas, G. P. , Diekman, B. O. , Krishnamurthy, J. , Hall, B. M. , Sorrentino, J. A. , Parker, J. S. , Sessions, G. A. , Gudkov, A. V. , & Sharpless, N. E. et al (2019) Cells exhibiting strong p16 INK4a promoter activation in vivo display features of senescence. Proceedings of the National Academy of Sciences of the United States of America, 116(7), 2603–2611. 10.1073/pnas.1818313116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. et al (2009). Expression of p16INK4a in peripheral blood T‐cells is a biomarker of human aging. Aging Cell, 8(4), 439–448. 10.1111/j.1474-9726.2009.00489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15(12), 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, N. , Beach, D. , & Gil, J. (2014). Ageing as developmental decay: insights from p16INK4a. Trends in Molecular Medicine, 20(12), 667–674. 10.1016/j.molmed.2014.09.008 [DOI] [PubMed] [Google Scholar]
- Martin‐Ruiz, C. , Williams‐Gray, C. H. , Yarnall, A. J. , Boucher, J. J. , Lawson, R. A. , Wijeyekoon, R. S. , Barker, R. A. , Kolenda, C. , Parker, C. , Burn, D. J. , Von Zglinicki, T. , & Saretzki, G. (2020). Senescence and Inflammatory Markers for Predicting Clinical Progression in Parkinson’s Disease: The ICICLE‐PD Study. Journal of Parkinson’s Disease, 10, 193–206. 10.3233/JPD-191724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, T. , Sankowski, R. , Staszewski, O. , & Prinz, M. (2020). Microglia Heterogeneity in the Single‐Cell Era. Cell Reports, 1271–1281. 10.1016/j.celrep.2020.01.010 [DOI] [PubMed] [Google Scholar]
- Melk, A. , Schmidt, B. M. W. , Takeuchi, O. , Sawitzki, B. , Rayner, D. C. , & Halloran, P. F. (2004). Expression of p16INK4a and other cell cycle regulator and senescence associated genes in aging human kidney. Kidney International, 65(2), 510–520. 10.1111/j.1523-1755.2004.00438.x [DOI] [PubMed] [Google Scholar]
- Neubrand, V. E. , Pedreño, M. , Caro, M. , Forte‐Lago, I. , Delgado, M. , & Gonzalez‐Rey, E. (2014). Mesenchymal stem cells induce the ramification of microglia via the small RhoGTPases Cdc42 and Rac1. Glia, 62(12), 1932–1942. 10.1002/glia.22714 [DOI] [PubMed] [Google Scholar]
- Nicaise, A. M. , Wagstaff, L. J. , Willis, C. M. , Paisie, C. , Chandok, H. , Robson, P. , Fossati, V. , Williams, A. , & Crocker, S. J. (2019). Cellular senescence in progenitor cells contributes to diminished remyelination potential in progressive multiple sclerosis. Proceedings of the National Academy of Sciences, 116(18), 9030–9039. 10.1073/pnas.1818348116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden, D. M. , & Godbout, J. P. (2013). Review: Microglia of the aged brain: Primed to be activated and resistant to regulation. Neuropathology and Applied Neurobiology, 19–34. 10.1111/j.1365-2990.2012.01306.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noseda, R. , Guerrero‐Valero, M. , Alberizzi, V. , Previtali, S. C. , Sherman, D. L. , Palmisano, M. , Huganir, R. L. , Nave, K.‐A. , Cuenda, A. , Feltri, M. L. , Brophy, P. J. , & Bolino, A. (2016). Kif13b Regulates PNS and CNS Myelination through the Dlg1 Scaffold. PLOS Biolog, 14(4), e1002440. 10.1371/journal.pbio.1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrodnik, M. , Evans, S. A. , Fielder, E. , Victorelli, S. , Kruger, P. , Salmonowicz, H. , Weigand, B. M. , Patel, A. D. , Pirtskhalava, T. , Inman, C. L. , Johnson, K. O. , Dickinson, S. L. , Rocha, A. , Schafer, M. J. , Zhu, Y. I. , Allison, D. B. , Zglinicki, T. , LeBrasseur, N. K. , Tchkonia, T. , … Jurk, D. (2021). Whole‐body senescent cell clearance alleviates age‐related brain inflammation and cognitive impairment in mice. Aging Cell, 20(2):13296. 10.1111/acel.13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, V. H. , & Holmes, C. (2014). Microglial priming in neurodegenerative disease. Nature Reviews Neurology, 10(4), 217–224. 10.1038/nrneurol.2014.38 [DOI] [PubMed] [Google Scholar]
- Peters, R. (2006). Ageing and the brain. Postgraduate Medical Journal, 82(964), 84–88. 10.1136/pgmj.2005.036665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, T. , Wang, K. , Cui, J. , He, Y. , & Yang, Z. (2016). Angiopoietin‐Like Protein 7 Promotes an Inflammatory Phenotype in RAW264.7 Macrophages Through the P38 MAPK Signaling Pathway. Inflammation, 39(3), 974–985. 10.1007/s10753-016-0324-4 [DOI] [PubMed] [Google Scholar]
- Raj, D. D. A. , Jaarsma, D. , Holtman, I. R. , Olah, M. , Ferreira, F. M. , Schaafsma, W. , Brouwer, N. , Meijer, M. M. , de Waard, M. C. , van der Pluijm, I. , Brandt, R. , Kreft, K. L. , Laman, J. D. , de Haan, G. , Biber, K. P. H. , Hoeijmakers, J. H. J. , Eggen, B. J. L. , & Boddeke, H. W. G. M. (2014). Priming of microglia in a DNA‐repair deficient model of accelerated aging. Neurobiology of Aging, 35(9), 2147–2160. 10.1016/j.neurobiolaging.2014.03.025 [DOI] [PubMed] [Google Scholar]
- Ramana, K. V. , Fadl, A. A. , Tammali, R. , Reddy, A. B. M. , Chopra, A. K. , & Srivastava, S. K. (2006). Aldose reductase mediates the lipopolysaccharide‐induced release of inflammatory mediators in RAW264.7 murine macrophages. Journal of Biological Chemistry, 281(44), 33019–33029. 10.1074/jbc.M603819200 [DOI] [PubMed] [Google Scholar]
- Robinson, M. D. , McCarthy, D. J. , & Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roney, K. E. , O'Connor, B. P. , Wen, H. , Holl, E. K. , Guthrie, E. H. , Davis, B. K. , Jones, S. W. , Jha, S. , Sharek, L. , Garcia‐Mata, R. , Bear, J. E. , & Ting, J.‐ P.‐Y. (2011). Plexin‐B2 Negatively Regulates Macrophage Motility, Rac, and Cdc42 Activation. PLoS One, 6(9), e24795– 10.1371/journal.pone.0024795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada, F. , Taniyama, Y. , Muratsu, J. , Otsu, R. , Shimizu, H. , Rakugi, H. , & Morishita, R. (2018). Source of Chronic Inflammation in Aging. Frontiers in Cardiovascular Medicine, 5, 12. 10.3389/fcvm.2018.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seya, T. , Oshiumi, H. , Sasai, M. , Akazawa, T. , & Matsumoto, M. (2005). TICAM‐1 and TICAM‐2: Toll‐like receptor adapters that participate in induction of type 1 interferons. International Journal of Biochemistry and Cell Biology, 524–529. 10.1016/j.biocel.2004.07.018 [DOI] [PubMed] [Google Scholar]
- Sierksma, A. , Lu, A. , Mancuso, R. , Fattorelli, N. , Thrupp, N. , Salta, E. , Zoco, J. , Blum, D. , Buée, L. , De Strooper, B. , & Fiers, M. (2020). Novel Alzheimer risk genes determine the microglia response to amyloid‐β but not to TAU pathology. EMBO Molecular Medicine, 12(3), 10.15252/emmm.201910606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic, M. R. , Ain, Q. , Bondeva, T. , Heller, R. , Schmeer, C. , & Witte, O. W. (2019). Phenotypic and functional differences between senescent and aged murine microglia. Neurobiology of Aging, 74, 56–69. 10.1016/j.neurobiolaging.2018.10.007 [DOI] [PubMed] [Google Scholar]
- Valério‐Gomes, B. , Guimarães, D. M. , Szczupak, D. , & Lent, R. (2018). ‘The absolute number of oligodendrocytes in the adult mouse brain. Frontiers in Neuroanatomy, 12, 10.3389/fnana.2018.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock, S. L. , Lopez, R. , & Klein, A. M. (2019). Scrublet: Computational Identification of Cell Doublets in Single‐Cell Transcriptomic Data. Cell Systems, 8(4), 281–291.e9. 10.1016/J.CELS.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss‐Coray, T. (2016). Ageing, neurodegeneration and brain rejuvenation. Nature, 180–186. 10.1038/nature20411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, S. , Gibilisco, L. , Kummer, M. , Biber, K. , Wachter, A. , & Woodbury, M. et al (2020). ABACUS: A flexible UMI counter that leverages intronic reads for single‐nucleus RNAseq analysis, bioRxiv. 10.1101/2020.11.13.381624 [DOI] [Google Scholar]
- Ximerakis, M. , Lipnick, S. L. , Innes, B. T. , Simmons, S. K. , Adiconis, X. , Dionne, D. , Mayweather, B. A. , Nguyen, L. , Niziolek, Z. , Ozek, C. , Butty, V. L. , Isserlin, R. , Buchanan, S. M. , Levine, S. S. , Regev, A. , Bader, G. D. , Levin, J. Z. , & Rubin, L. L. (2019). Single‐cell transcriptomic profiling of the aging mouse brain. Nature Neuroscience, 22(10), 1696–1708. 10.1038/s41593-019-0491-3 [DOI] [PubMed] [Google Scholar]
- Xu, M. , Pirtskhalava, T. , Farr, J. N. , Weigand, B. M. , Palmer, A. K. , Weivoda, M. M. , Inman, C. L. , Ogrodnik, M. B. , Hachfeld, C. M. , Fraser, D. G. , Onken, J. L. , Johnson, K. O. , Verzosa, G. C. , Langhi, L. G. P. , Weigl, M. , Giorgadze, N. , LeBrasseur, N. K. , Miller, J. D. , Jurk, D. , … Kirkland, J. L. (2018). Senolytics improve physical function and increase lifespan in old age. Nature Medicine, 24(8), 1246–1256. 10.1038/s41591-018-0092-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabluchanskiy, A. , Tarantini, S. , Balasubramanian, P. , Kiss, T. , Csipo, T. , Fülöp, G. A. , Lipecz, A. , Ahire, C. , DelFavero, J. , Nyul‐Toth, A. , Sonntag, W. E. , Schwartzman, M. L. , Campisi, J. , Csiszar, A. , & Ungvari, Z. (2020). Pharmacological or genetic depletion of senescent astrocytes prevents whole brain irradiation–induced impairment of neurovascular coupling responses protecting cognitive function in mice. GeroScience, 42(2), 409–428. 10.1007/s11357-020-00154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefzadeh, M. J. , Zhao, J. , Bukata, C. , Wade, E. A. , McGowan, S. J. , Angelini, L. A. , Bank, M. P. , Gurkar, A. U. , McGuckian, C. A. , Calubag, M. F. , Kato, J. I. , Burd, C. E. , Robbins, P. D. , & Niedernhofer, L. J. (2020). Tissue specificity of senescent cell accumulation during physiologic and accelerated aging of mice. Aging Cell, 19(3), 10.1111/acel.13094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Velmeshev, D. , Hashimoto, K. , Huang, Y.‐H. , Hofmann, J. W. , Shi, X. , Chen, J. , Leidal, A. M. , Dishart, J. G. , Cahill, M. K. , Kelley, K. W. , Liddelow, S. A. , Seeley, W. W. , Miller, B. L. , Walther, T. C. , Farese, R. V. , Taylor, J. P. , Ullian, E. M. , Huang, B. O. , … Huang, E. J. (2020). Neurotoxic microglia promote TDP‐43 proteinopathy in progranulin deficiency. Nature, 588(7838), 459–465. 10.1038/s41586-020-2709-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Kishimoto, Y. , Grammatikakis, I. , Gottimukkala, K. , Cutler, R. G. , Zhang, S. , Abdelmohsen, K. , Bohr, V. A. , Misra Sen, J. , Gorospe, M. , & Mattson, M. P. (2019). Senolytic therapy alleviates Aβ‐associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nature Neuroscience, 22(5), 719–728. 10.1038/s41593-019-0372-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1‐S3
Table S1‐S7
Data Availability Statement
RNAseq data are deposited in the database GEO (www.ncbi.nih.gov/geo/) with identifier GSE151459. All the data presented here are available from the corresponding authors upon reasonable request.


