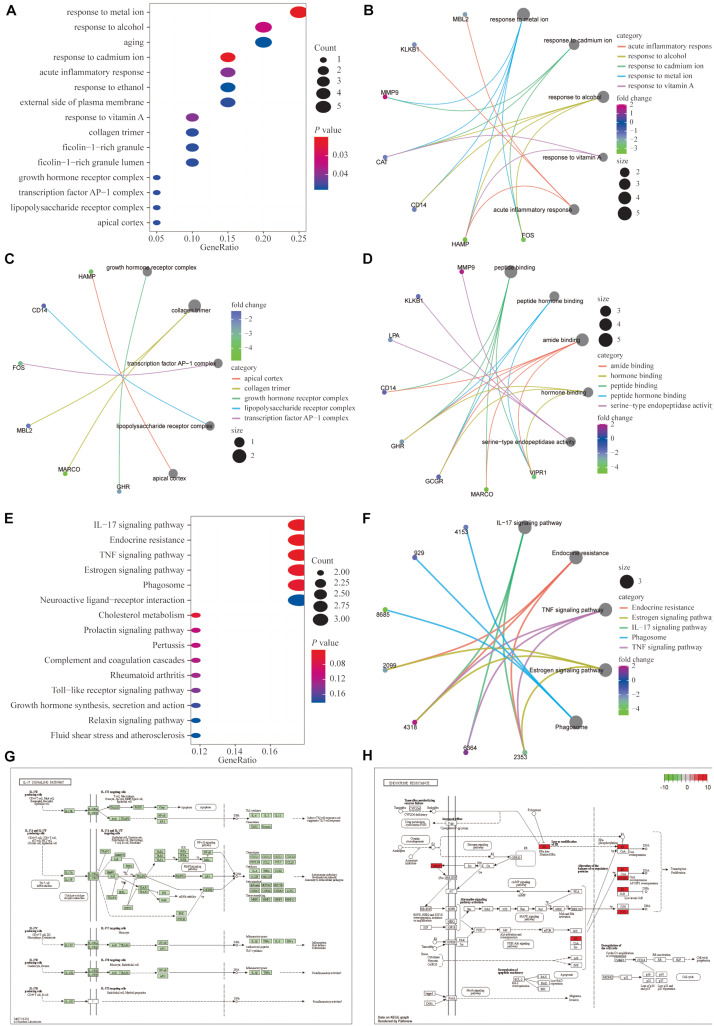
FIGURE 2.
Functional enrichment analysis of the 20 differentially expressed IRGs. (A–D) Gene ontology (GO) analysis showed that the differentially expressed genes (DEGs) were closely associated with biological processes such as responses to metal ion, response to cadmium ion, growth hormone receptor complex, collagen trimer, and peptide binding. (E,F) Kyoto encyclopedia of genes and genomes (KEGG) functional analysis suggested that differentially expressed immune genes mainly affected the IL-17, endocrine resistance, TNF and estrogen signaling pathways. (G,H) Detailed demonstrations of the IL-17 and endocrine resistance pathways.