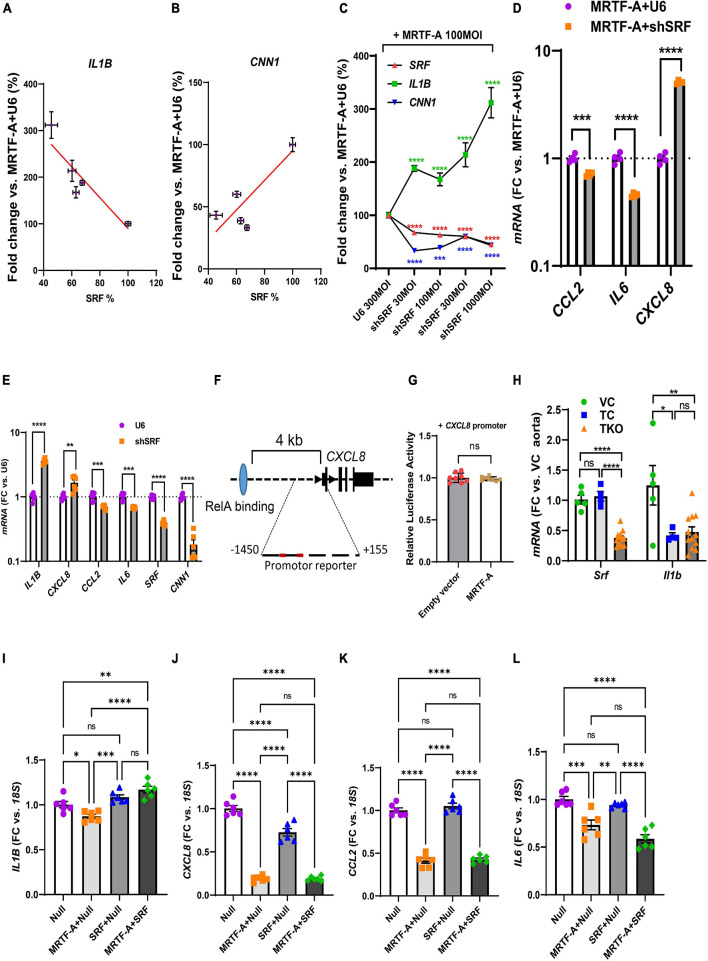
FIGURE 6.
SRF is important for regulation of IL1B and CXCL8. (A–C) Human coronary SMCs were transduced with MRTF-A along with different titers of Ad-h-shSRF for knockdown of SRF. After harvesting the cells and isolation of RNA, SRF, IL1B, and CNN1 were assayed using RT-qPCR. In (A,B), IL1B and CNN1 were plotted vs. the relative SRF level in the respective samples. In (C), SRF, IL1B, and CNN1 were plotted vs. the titer (multiplicity of infection, MOI) of the short hairpin virus. (D) Shows the remainder of the inflammatory mediators in control vs. SRF-depleted cells (1000 MOI). (E) Is like (D), except that SRF silencing was done without simultaneous overexpression of MRTF-A. (F) Shows the gene locus for human CXCL8 and binding of RelA (light blue oval) 4 kb upstream of the transcription start site. The proximal promoter contained two DNA sequences with two deviations each from the classical SRF-binding sequence. When testing this sequence in a reporter assay, no suppression by MRTF-A was however, seen (G). In (H), Srf and Il1b were assayed in the aorta from smooth muscle specific and inducible Srf knockout mice. Because knockout was induced by tamoxifen, two control groups were included in addition to the knockout group. Vehicle controls (VC) are Cre-positive mice injected with sunflower oil, whereas tamoxifen controls (TC) are Cre-negative mice injected with tamoxifen. Knockouts (TKO) are Cre-positive mice injected with tamoxifen. All mice are homozygous for the floxed Srf allele. (I–L) Show the effect of overexpression of SRF in the absence and presence of MRTF-A. SRF was capable of suppressing CXCL8 on its own but was without effect on CCL2 and IL6. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.