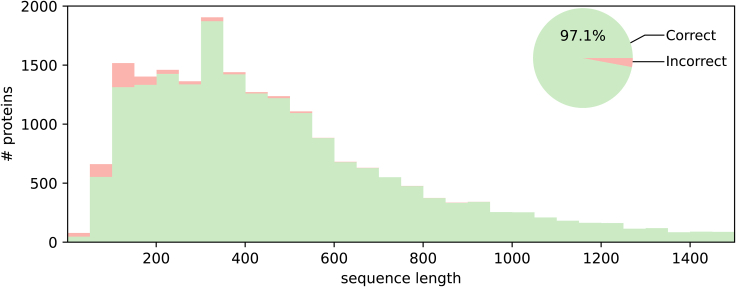
Figure 2.
Simulated fingerprint identification accuracy assuming low-noise conditions
Cumulative histogram of correct and incorrect classifications of simulated chop-n-drop protein fingerprints for all human proteome constituents, assuming low noise parameters; resolution r = 5 Da, capture rate C = 0.99 and proteasome cleaving efficiency ep = 0.99. Numbers shown are distributed over sequence length (bars) and relative to the total number of proteins (pie chart). Alignment examples and sequence identity distribution for erroneous alignments are shown in Figures S3 and S4, respectively. Results assuming charge-dependent fragment capture are shown in Figure S5A.