Figure 4. GRASP55 interacts with GCS and LCS.
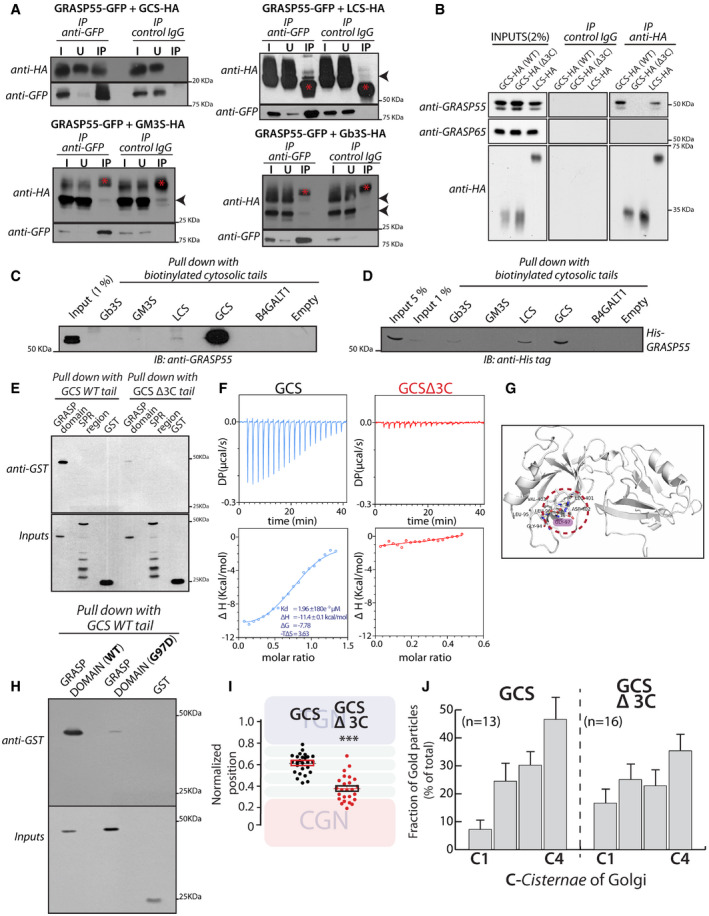
- HeLa cells co‐transfected with indicated HA‐tagged enzymes (GCS, LCS, GM3S, and Gb3S) and GRASP55‐GFP were lysed, immunoprecipitated with anti‐GFP antibody or control IgG, and were analyzed by Western blotting for interaction by immunoblotting (IB) with anti‐HA antibody. I represents 5% on the input lysate, U 5% of the unbound fraction, and IP the immunoprecipitate. Red asterisks indicate IgG bands, and arrow heads indicate the expected position of HA‐tagged enzymes
- WI‐26 fibroblasts transfected with the indicated HA‐tagged enzymes were lysed, immunoprecipitated with anti‐HA antibody or control IgG, and were analyzed by Western blotting for interaction by immunoblotting with the indicated antibodies.
- Chemically synthesized biotinylated peptides corresponding to cytosolic portions of glycosylation enzymes were bound to avidin beads and were used to pull down interactors from HeLa cell lysates and subjected to immunoblotting with anti‐GRASP55 antibody.
- The interaction of chemically synthesized biotinylated peptides, corresponding to cytosolic portions of glycosylation enzymes, with purified His‐tagged full‐length GRASP55 and their interaction was monitored by pulling down the biotinylated peptides bound to avidin beads followed by Western blotting with anti‐His tag antibody.
- Chemically synthesized biotinylated peptides corresponding to cytosolic portions of GCS (WT and Δ3C) and indicated purified GST‐tagged GRASP domain or SPR region of GRASP55 were incubated together, and their interaction was monitored by pulling down the biotinylated peptides with avidin beads followed by Western blotting with an anti‐GST tag antibody.
- ITC profile, representative of at least two independent experiments, for biotinylated GCS and GCS Δ3C cytosolic tails with recombinant GRASP55.
- The molecular basis of interaction between GRASP55 and ceramide glucosyltransferase C‐terminal peptide is studied by building a model of GRASP55:GCS peptide structure in the absence of the complex crystal structure. The carboxylate group of Leu of “LDV” motif retains conserved hydrogen bonds with the backbone of 95LLGV98motif of GRASP55. Gly97 residue which crucial to GRASP:GCS interaction is highlighted (pink).
- The interactions of chemically synthesized biotinylated peptides corresponding to cytosolic portions of GCS (WT) with the indicated purified GST‐tagged GRASP domain (WT) or GRASP domain (G97D) were monitored by pulling down the biotinylated peptides with avidin beads followed by Western blotting with an anti‐GST tag antibody.
- HeLa cells were transfected with either WT GCS or GCS Δ3C, treated with nocodazole (33 µM) for 3 h and labeled for enzymes, GM130, and TGN46 (to mark CGN and TGN respectively). Line scan analysis was performed as in Fig 3A and B, and the relative position of enzymes was quantitated and plotted. The data are mean ± SD (n = 30) representative of two experiments. **P < 0.01, ***P < 0.001(Student’s t‐test) and ns signifies not statistically significant.
- HeLa cells were transfected with either WT GCS or GCS Δ3C for 16 h and processed for cryoimmunolabeling. Distribution of indicated enzymes across the Golgi stack was quantified and represented as fraction of Gold particles in each cisterna (n indicated in the graph; data are Mean ± SEM).
Source data are available online for this figure.
