FIGURE 2.
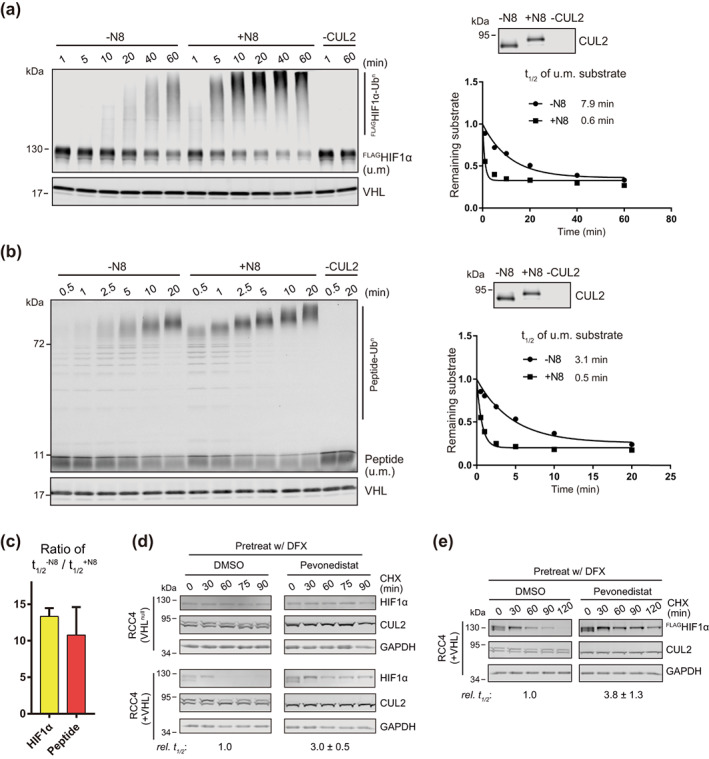
Neddylation is required for efficient CRL2VHL‐dependent degradation of HIF1α in vitro and in RCC4 (+VHL) cells. (a,b) Full‐length FLAGHIF1α purified from RCC4 (VHL null) cells stably expressing FLAGHIF1α (a) or HIF1α degron peptide (b) were ubiquitinated in vitro for indicated time with or without CUL2 neddylation. Samples without CUL2 served as negative controls. Intensities of unmodified (u.m.) FLAGHIF1α or degron peptide bands were normalized to intensities of VHL bands for regression analyses. WB of CUL2 from each group and regression curves of remaining unmodified substrates were shown on the right. (c) Average relative changes of substrate t 1/2 by neddylation from experiments in (a,b). Error bars: range of values, n = 2. (D) RCC4 (VHL null) and RCC4 (+VHL) cells were pretreated with DFX for 3 h and pevonedistat or equivalent volume of DMSO for 1 h. After washout of inhibitors, cells were treated by CHX with or without pevonedistat for indicated time and were collected for WB analyses. (e) Same assays as performed in (d) with FLAGHIF1α stably expressed in RCC4 (+VHL) cells. In (d,e), relative t 1/2 was shown as average ± range of values (n = 2)
