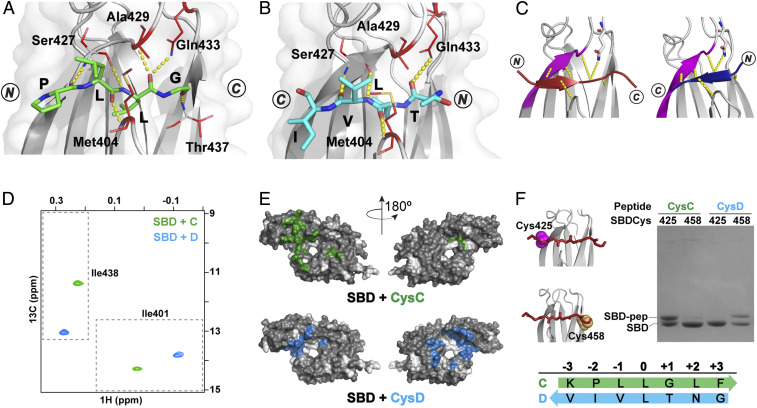
Fig. 4.
Peptides containing proPhoA sites C and D bind to the DnaK SBD in opposite orientations. (A and B) Crystal structures (top view) of the SBD (gray) in complex with proPhoA peptide C (A), which binds in an N to C orientation (green), and Dsh (B), which binds in a C to N orientation (blue). Residues 507 to 603 are not shown. The side chains of SBD residues contacting the peptide are in red sticks, and hydrogen bonds between the peptide backbone and the SBD are in yellow. (C) Schematic showing that the bound peptide interacts with the β3-strand of the SBD (magenta) in a parallel or antiparallel strand–strand arrangement for the N to C [Left; peptide in maroon, PDB ID code 1DKZ (11)] or the C to N binding mode [Right; peptide in blue, PDB ID code 4EZY (12)]. (D) The Ile401 and Ile438 region of 1H-13C-HMQC spectra of ILV-13CH3–labeled DnaK SBD in the presence of unlabeled peptides C (green) and D (blue). (Peptides and SBD are at 40 μM.) (E) PRE NMR of SBD complexes with CysC and CysD peptides. SBD residues significantly broadened in the presence of spin-labeled peptides are mapped on the SBD structure (in green, CysC; in blue, CysD). Residues for which no data are available are shown in white. (F) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of SBDCys425 and SBDCys458 (cysteine residues in magenta and yellow spheres in the structures in Left) cross-linked to CysC and CysD peptides.