Fig. 7.
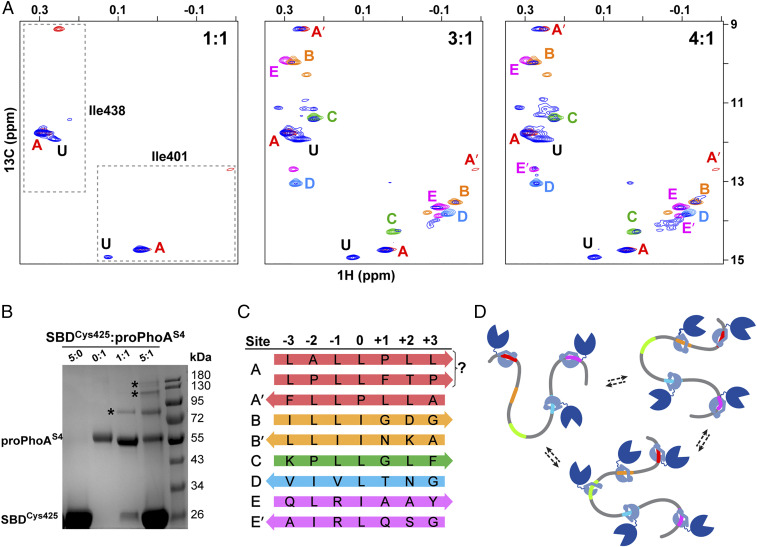
From peptide to protein, the DnaK SBD binds multiple sites on proPhoA with differing affinities, largely consistent with studies of peptides, and one previously unidentified high-affinity site. (A) Titration of unlabeled proPhoAS4 with increasing amounts of ILV-13CH3 DnaK SBD. The Ile401 and Ile438 resonances of the SBD in complex with proPhoAS4 are shown in blue, and resonances for complexes with the proPhoA peptides A to E are color coded as in Fig. 3. At a 1:1 ratio of SBD to proPhoAS4, the resonances for the protein complex coincide with those for peptide A (red), and an additional resonance is seen (labeled U) that does not correspond to any of the peptides tested. At a 3:1 ratio of SBD to proPhoAS4, additional resonances appear that overlay on those for the SBD complexes with peptides C and D and major species for complexes with peptides B and E. Finally, at a 4:1 ratio of SBD to proPhoAS4, all the resonances seen for the peptides appear. (B) SDS-PAGE of SBDCys425 cross-linked to proPhoAS4 using Sulfo-GMBS. Addition of the cross-linker to SBD or proPhoAS4 alone does not result in higher–molecular mass species (lanes 1 and 2). At a 1:1 ratio of SBD to proPhoAS4, a 1:1 complex is observed (lane 3), while at a 5:1 ratio of SBD to proPhoAS4, complexes with multiple SBD bound to one proPhoA molecule are seen. (Controls are shown in SI Appendix, Fig. S13.) (C) Summary of DnaK SBD binding sites on proPhoAS4, their occupancy of SBD binding pockets, and their orientation. The prime symbol indicates an alternative minor binding mode. (D) Cartoon depicting DnaK binding to proPhoA. Site A, which has the highest binding affinity to DnaK, is occupied by the chaperone at the lowest chaperone to substrate ratios and remains bound as the chaperone to substrate ratio increases. Other binding sites on proPhoA are occupied based largely on the affinities they manifest as peptides but in a dynamic manner, with several or all of them occupied by chaperone simultaneously.