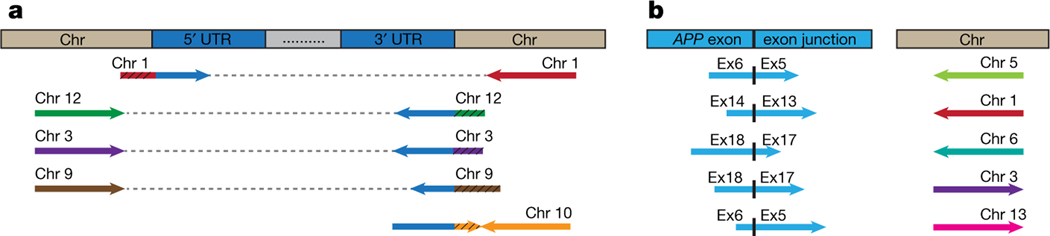
Fig. 1 |. Identification of novel APP insertion sites in the human genome.
a, Clipped reads spanning APP UTRs and novel chromosomal insertion sites were identified. The paired mate-reads of the clipped reads (black hatching) uniquely mapped to the same chromosomes. b, Discordant read-pairs were identified where one read spanned an APP exon–exon junction and the corresponding mate-read mapped to a novel chromosome. Each chromosome has a unique colour. Arrowhead direction represents the read orientation after mapping to the human reference genome. Arrows oriented in the same direction support sequence inversions. See detailed sequence and alignment information in Supplementary Table 1.