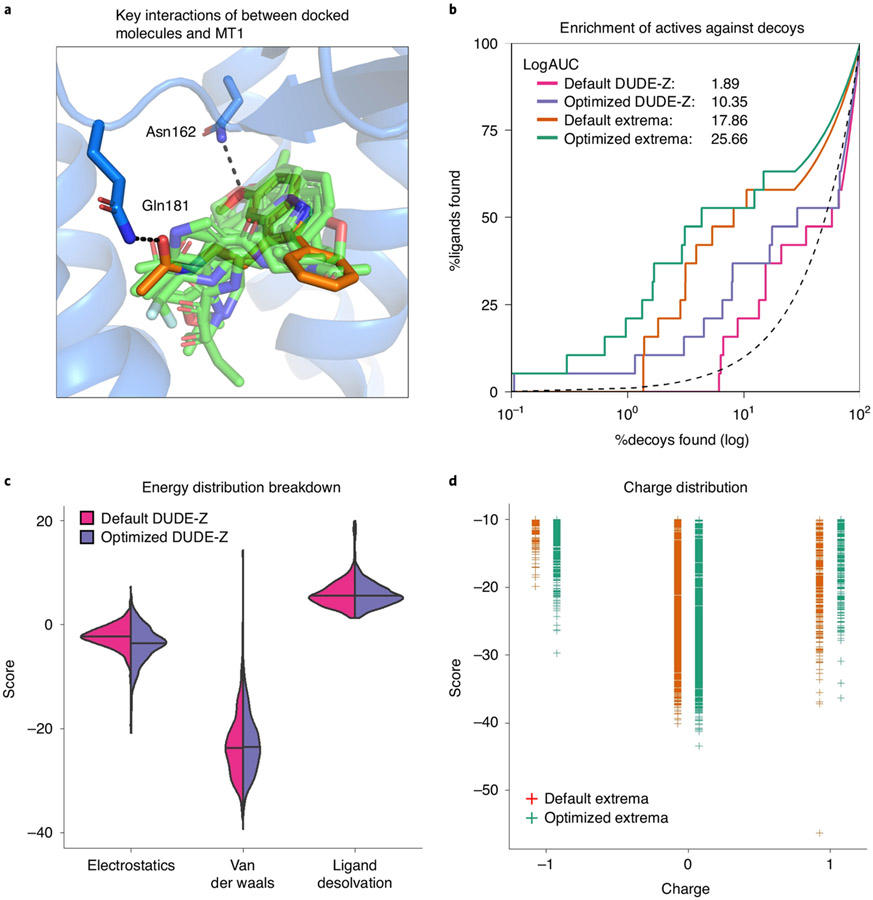
Fig. 6 ∣. Controls for docking optimization.
a, The receptor (blue) is shown with the crystallized ligand (orange). Docked control actives are shown in green and yield similar poses and interactions as the crystal ligand. The two residues, Asn162 and Gln181, that have their dipoles artificially increased (‘polarized’) to enhance the weight of polar interactions are shown hydrogen bonding to the crystal ligand. b, A log-transformed ROC plot is shown comparing the rate of identifying ligands versus decoys. A random selection would follow the dashed black line. The area under this dashed line is subtracted from the values reported for LogAUC such that a curve above the line would have a positive LogAUC, a curve below the line would have a negative LogAUC, and a curve following the dashed line would yield a LogAUC value of zero. Shown are the curves for the default settings and optimized settings for either the DUDE-Z control set and the Extrema control set. In both cases, the overall LogAUC value increases and the early enrichment improves. c, The energy distribution breakdown shows the individual score terms for each scored molecule in the docked setup. Based on this breakdown, it is clear that VDW interactions primarily drive ligand recognition. However, in the optimized setup in which electrostatic spheres with a radius of 1.9 that extend the dielectric boundary are used, the electrostatic score term shifts to more negative values. The desolvation spheres at the dielectric boundary in the optimized setup, with a radius of 0.1, have only minor effects on the ligand desolvation score term. d, In the Extrema challenge, the top-ranking ligands are plotted by their charge and DOCK score. In the Default settings, there is a preference for neutral ligands followed closely by monocations. The Optimized settings enhance the preference for neutrals.