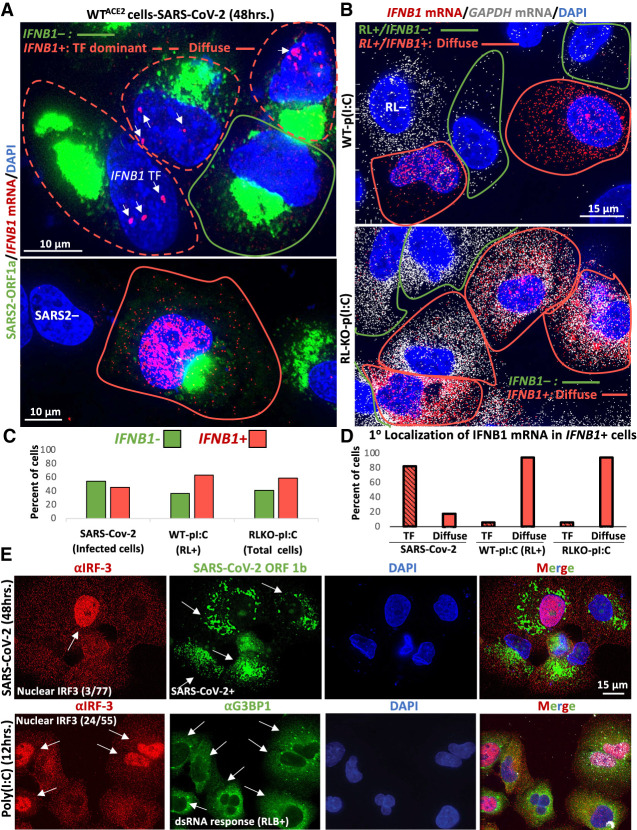
FIGURE 5.
IFN mRNAs are retained at the site of transcription during SARS-CoV-2 infection. (A) smFISH for IFNB1 mRNA and SARS-CoV-2 ORF1a 48 h post-infection. Two fields of view are shown. In the top image, the cell boundary of SARS-CoV-2-positive cells that stain for IFNB1 are demarcated by red line, whereas IFNB1 mRNA-negative cells are demarcated by green line. Cells that contain IFNB1 transcription site foci (TF) but lack abundant disseminated IFNB1 mRNA are demarcated by dashed red line. The lower image shows a SARS-CoV-2-infected cell that contains abundant and diffuse IFNB1 mRNA in the nucleus and cytoplasm. Cells that do not stain for SARS-CoV-2 are labeled SARS2-. (B) smFISH for IFNB1 mRNA and GAPDH mRNA 16 h post-poly(I:C) transfection in WT and RL-KO A549 cells. In WT cells, 12% do not activate RNase L (RL−). Of the 88% of cells that activate RNase L, 63% (55% of total cells) also induce abundant and disseminated IFNB1 mRNA (red line), whereas 37% of RL+ cells do not induce IFNB1 (green line). Fifty-nine percent of RL-KO cells induce abundant disseminated IFNB1 mRNA (red line), whereas 41% do not (green line). (C) Histograms quantifying the percent of SARS-CoV-2 infected cells, poly(I:C)-transfected WT cells that activate RNase L (GAPDH mRNA-negative cells), and poly(I:C)-transfected RL-KO cells that induce IFNB1, as represented in A and B. (D) Histograms quantifying the percent of IFNB1-positive cells in which IFNB1 smFISH staining is predominantly localized to IFNB1 transcription site foci (TF) or diffuse. (E) Immunofluorescence assay for IRF3 translocation from the cytoplasm to the nucleus in response to either SARS-CoV-2 infection (48 h p.i.; MOI = 5) or poly(I:C) lipofection (12 h). The fraction of cells displaying robust nuclear IRF3 staining is shown in the IRF3 images. For SARS-CoV-2 infection, smFISH for SARS-CoV-2 ORF-1b was used to identify infected cells indicated by arrows. For cells undergoing dsRNA response to poly(I:C), G3BP1 immunofluorescence was used to identify RNase L-dependent bodies (RLBs), as indicated by white arrows.