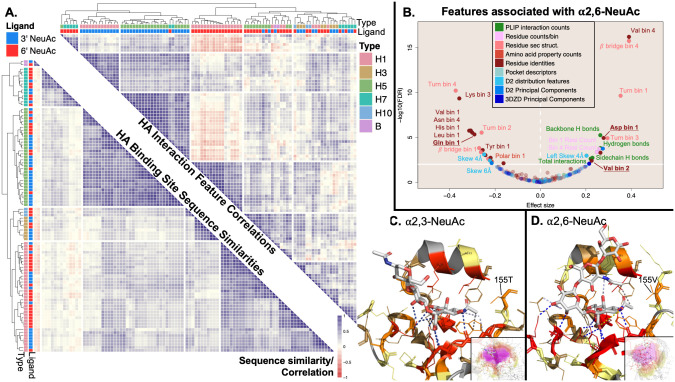
Fig 6. Focused analysis of influenza HA binding sites reveals significant and discriminative features associated with binding of human-like sialoglycans over avian-like sialogylcans.
Clustering HA interactions by significant interaction features discriminates those recognizing 3’ vs. 6’ NeuAc terminal glycans, while clustering by interaction sequence simply recapitulates influenza strain. Panel A shows that clustering of the 96 HA-3’/6’ αNeuAc interactions using correlations from the 35 significantly associated features allows for much cleaner grouping of interactions by ligand-type (upper-right-triangular similarity matrix) compared to interaction clustering using the alignment of the sequence of binding site residues leading to perfect grouping of interactions by influenza strain and HA subtype in the lower-left-triangular similarity matrix. Comparisons between hemagglutinin structures with 6’ αNeuAc-terminal glycans versus 3’ αNeuAc-terminal glycans reveal 35 features that are significantly associated with the presence of 6’ αNeuAc-terminal glycans (panel B) displayed in the same manner as in Fig 2 with points discussed directly in the text bolded and underlined. These features are found in representative interaction structures between the respective glycans and HA proteins in panels C & D. In panel A, the upper-right-triangular matrix was constructed by calculating the pairwise Pearson correlations for all interactions using the scaled values of the 35 significant interaction features. The lower-left-triangular similarity matrix was constructed from sequence similarity scores using Needleman-Wunch to align binding site sequences with the BLOSUM62 substitution matrix. In panel B, significance and effect size were determined by a Wilcoxon-Mann-Whitney test weighted by influenza strain/hemagglutinin subtype and UniProt ID, with a significance threshold of q < 0.01 (solid horizontal white line) by the Benjamini-Hochberg Procedure. Panel C shows HA from H1N1 (Puerto Rico/8/1934) (dual specificity) complexed with an avian sialopentasccharide, although only the three terminal sugars were resolved (PDB ID: 1RVX). Panel D shows HA from H1N1 (California/4/09) in complex with a human sialopentasccharide (PDB ID: 3UBE). Both panels C and D use the same color scheme for lectins, PLIP interactions, and glycans as in Fig 5.