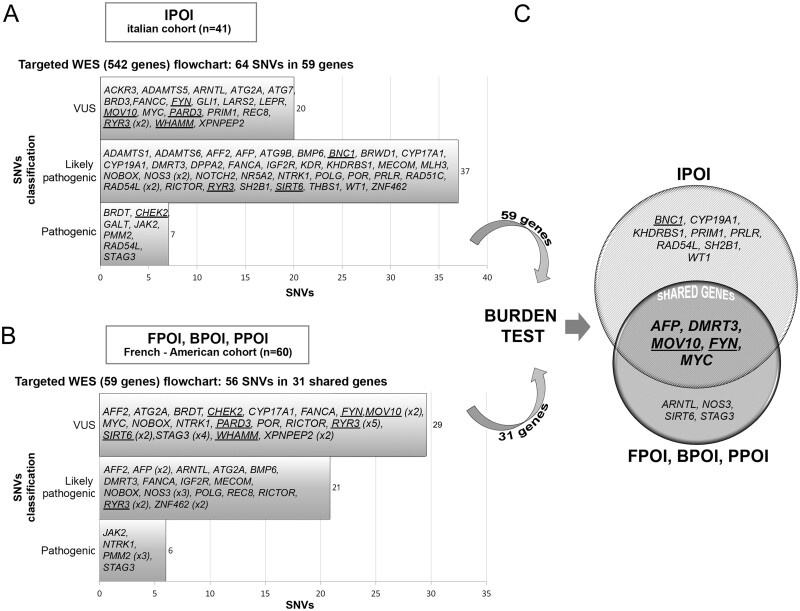
Figure 2.
Flowchart of the results. (A) The Italian cohort (IPOI) of 41 individuals was analysed by means of a targeted (542 genes) WES. The upper left horizontal bars show the number of variants identified in IPOI according to the ACMG/AMP classification. The genes bearing the selected alterations (n = 59) are listed inside the bars. (B) The same approach was applied to a second cohort of French and American patients (FPOI, BPOI, PPOI) starting from the 59 genes identified in the Italian cohort. The bottom left histograms show the number of variants identified in FPOI, BPOI and PPOI according to the ACMG/AMP classification. The genes harbouring the selected alterations and shared with the Italian cohort (n = 31) are listed inside the bars. (C) Burden test analysis performed on the genes identified through targeted-WES in both cohorts of patients revealed significant genes in each cohort, which are shown in the Venn diagram. The shared significant genes are in bold and upper case letters inside the intersection. Genes identified by array-CGH analysis (Bestetti et al., 2019) and the genes encoding for interactors of the main array-CGH candidates, are shown underlined. ACMG/AMP, American College of Medical Genetics and Genomics; CGH comparative genomic hybridisation; POI, primary ovarian insufficiency; WES, whole exome sequencing.