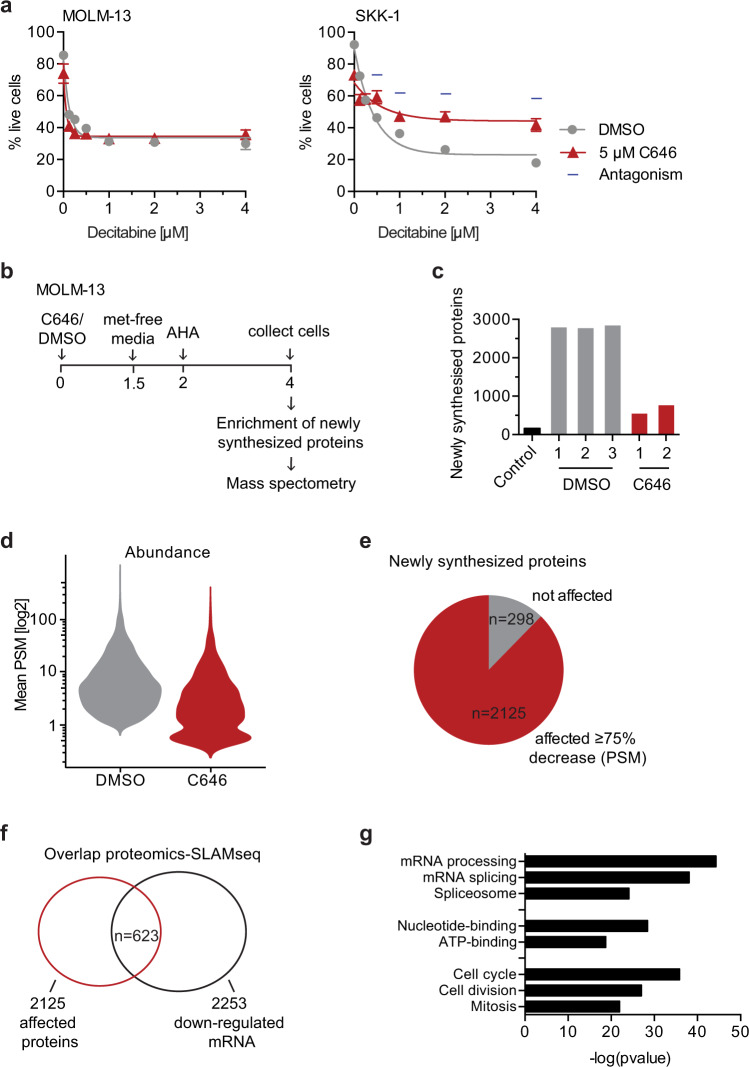
Fig. 4. CBP/p300 inhibition reduces protein synthesis but does not synergize with decitabine.
a Percentage of live MOLM-13 or SKK-1 cells after 4 days of treatment with indicated concentrations of AZA in combination with 5 µM decitabine or DMSO (0.05%) as vehicle control. Data represent the mean ± SEM of four independent experiments. Statistical analysis was performed using two-way ANOVA. Calculating the combination index (CI) indicated antagonism (CI > 1). b Schematic workflow of the detection of nascent proteins. MOLM-13 cells were treated with 10 µM C646 or 0.1% DMSO, the media was changed to methionine (met)-free media after 1.5 h, metabolic AHA label was added for 2 h and cells were collected after a total of 4 h of inhibitor treatment. Newly synthesized proteins were affinity purified following the ClickIT protocol. c The number of detected newly synthesized proteins in untreated and treated samples is plotted. An unlabelled sample served as negative control. d Violin plot presenting the mean peptide spectrum matches (PSM) of all detected proteins. e Pie chart indicating that the majority of detected proteins was affected as defined by a reduction in PSM of 75% or more. f Overlap between significantly decreased newly synthesized proteins (≥75% reduction in PSM) determined by ClickIT enrichment and significantly downregulated nascent mRNA (log2FC ≤ −1, FDR ≤ 0.1) determined by SLAM-seq. g Ontology analysis of significantly reduced proteins (≤75% reduction compared to untreated samples). Statistical analysis was performed by Fisher’s exact test. Source data are provided as a Source Data file and in Supplementary Data 6 and 7.