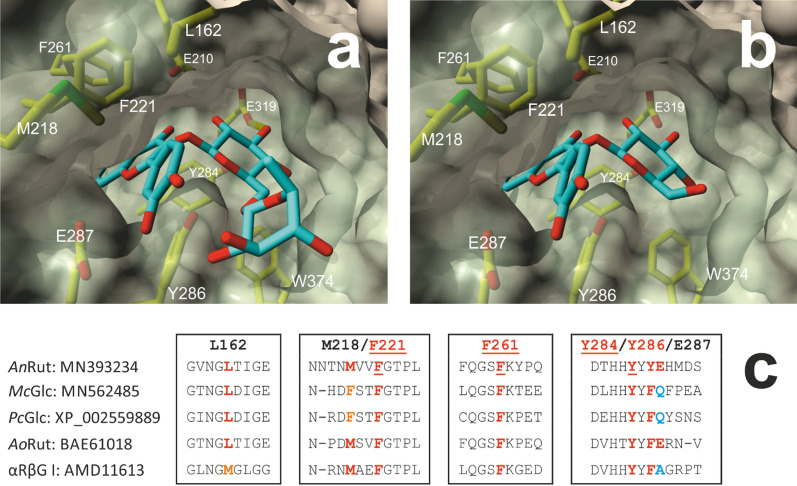
Fig. 2.
View of the main entrance to the active site of AnRut with bound substrate 1 or 2 and multiple sequence alignment of GH5-23 member enzymes. a Molecular model of 2 docked to the active site with a free energy of binding of –10.2 kcal mol–1. b Molecular model of docked 1 with a free energy of binding of –9.3 kcal mol–1. Residues that interact with the bound substrates are labeled and shown in stick representation. These include in particular the aromatic sidechains of F221, F261, Y284 and Y286, which interact with the aglycones via many hydrophobic and π–π interactions (Additional file 1: Tables S2–S5). In addition, hydrophobic interactions between the aglycones and the sidechains of L162, M218 and E287 were also found. The catalytic nucleophile E319 and the acid/base catalyst E210 are shown as well. The most favorable binding poses of 1 and 2 are shown. The double ring moiety of quercetin (rings A and C; see Fig. 1) is buried in the side tunnel and only incompletely visible. The above-mentioned residues were set flexible during the simulation. The AnRut-based residues are covered with a partially transparent molecular protein surface. c Multiple sequence alignment of GH5-23 member enzymes. Only segments around residues that—according to the molecular docking results—are involved in hydrophobic and π-π interactions between the quercetin moiety of 1 or 2 and the + 1 subsite of AnRut are shown. The aromatic residues F221, F261 and Y284 are fully conserved. The residues at the positions 162, 218 and 286 are partially conserved. The numbering refers to the sequence of AnRut (MN393234) (Pachl et al. 2020). The following sequences were aligned: MN562485 (McGlc) and XP_002559889 (PcGlc) (Kotik et al. 2021), BAE61018 (AoRut; Makabe et al. 2021), AMD11613 (αRβG I; Weiz et al. 2019)