Abstract
The clinical phenotypes of nonalcoholic fatty liver disease (NAFLD) encompass from simple steatosis to nonalcoholic steatohepatitis (NASH) with varying degrees of fibrosis or cirrhosis. Liver biopsy has been the standard to diagnose NASH. However, there has been strong need for precise and accurate noninvasive tests because of invasiveness and sampling variability of biopsy. Metabolomics has drawn attention as a promising diagnostic methodology in the field of NAFLD, particularly to unravel metabolic alterations which plays relevant roles in the progression of NASH. There have been numerous metabolomics researches to find new biomarker of NASH in the last decade, fueled by the recent advances in the metabolomics methodology. This review briefly covers recent research advances on the lipidomics, amino acids and bile acid metabolomics regarding continuing attempts to discover relevant biomarkers for NASH.
Keywords: Lipids, Bile acids and salts, Amino acids, Fatty liver, Phenotype
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is currently the most frequent cause of chronic liver disease worldwide, affecting approximately 25% of entire adult population [1]. NAFLD is a spectrum of liver diseases, ranging from simple hepatic steatosis, or nonalcoholic fatty liver (NAFL), to nonalcoholic steatohepatitis (NASH) with varying degrees of fibrosis, or even cirrhosis [2]. In particular, NASH is characterized by the histologic evidence of inflammation with hepatocyte injury (e.g., ballooning) with or without any fibrosis, on top of hepatic steatosis. Unlike the benign clinical course of NAFL, NASH can progress to cirrhosis, liver failure as well as development of hepatocellular carcinoma. Liver biopsy has been the reference standard for the diagnosis of NASH, i.e., the aggressive phenotype of NAFLD [3]. However, because of its shortcomings such as invasiveness and interobserver variability which make biopsy less practical for clinical diagnostics or disease monitoring, many noninvasive tests have been developed to identify patients with NASH or with higher risk of NASH. Up to now, most available noninvasive tests are proven to be effective for identifying advanced fibrosis rather than the presence of NASH [4].
Metabolomics has been extensively utilized as a promising investigational method for small molecules and metabolic products, including amino acids, fatty acids, and carbohydrates, in the field of NAFLD research [5]. Many researchers have investigated pathophysiologic role of the metabolome in animal models or patients with NAFLD. This review covers recent advances in metabolomics as a diagnostic tool for NASH vs. NAFL, particularly focusing on human studies of the lipidomics, amino acids, and bile acid metabolomics.
DIAGNOSIS OF NAFLD PHENOTYPES USING LIPIDOMICS
Recent researches have endeavored to develop diagnostic models using metabolomics, e.g., lipidomics, with or without other laboratory or clinical parameters. Aims of such models mainly focused on determination of NAFLD phenotypes, such as presence or absence of fibrosis, and differentiation between NAFL vs. NASH. Technical advances also helped those analysis especially in mass spectrometry (MS) methods and standardization of reporting identified lipids.
Several diagnostic algorithms to figure out NAFLD phenotypes have been developed based on relevant parameters from targeted or untargeted approaches, followed by creating algorithms using logistic regression. In Zhou et al.’s study [6], a prediction model was developed and validated, consisting of aspartate aminotransferase (AST), insulin, and patatin-like phospholipase domain-containing-3 (PNPLA3) genotype, which achieved an area under the receiver operating characteristic (AUROC) of 0.778 in terms of identification of NASH (NASH Clin Score). They further incorporated additional MS-based lipids and metabolites markers in the score, i.e., glutamate, glycine, isoleucine, lysophosphatidylcholine (16:0), and phosphatidylethanolamine (40:6). The latter model, named NASH ClinLipMet Score, showed improved differentiation of NASH vs. non-NASH with an AUROC of 0.866, with a sensitivity of 85% and a specificity of 72%, respectively [6]. In another study by Mayo et al. [7], a diagnostic algorithm was developed based on 20 kinds of serum triglyceride species from a large cohort of biopsy-proven patients (derivation cohort of 467 patients [90 control, 246 NAFL, 131 NASH] and validation cohort of 192 patients [seven control, 109 NAFL, 76 NASH]), which differentiated NASH from NAFL with an AUROC of 0.79, 70% sensitivity and 81% specificity, respectively.
In an earlier study by Barr et al. [8], a body mass index-dependent serum metabolic profile reliably identified NASH from NAFL, based on which a logistic regression algorithm (OWLiver test) was developed using 20 triglycerides to distinguish NASH and simple steatosis. However, the performance of this test was not satisfactory in patients with type 2 diabetes mellitus (T2DM) and NAFLD (AUROC, 0.69), whereas the test was originally derived in patients without T2DM [9]. The discrepancy observed in those two studies suggest that prediction of NAFLD phenotypes is affected significantly by the characteristics of the patient population where those models were derived.
Fibrosis stage is a well-known prognostic factor in terms of morbidity and mortality in NAFLD patients [10,11]. A study using untargeted metabolite profiling by Caussy et al. [12] found 10 relevant metabolites to predict the presence of advanced fibrosis, i.e., eight lipids (5alpha-androstan-3beta monosulfate, pregnanediol3-glucuronide, androsterone sulfate, epiandrosterone sulfate, palmitoleate, dehydroisoandrosterone sulfate, 5alphaandrostan-3beta disulfate, glycocholate), one amino acid (taurine), and one carbohydrate (fucose). The AUROC of the serum metabolite panel was 0.94, outperforming FIB-4 and NAFLD Fibrosis Score, and maintaining its diagnostic accuracy in the independent validation cohorts. The same research group demonstrated the association between plasma eicosanoids and liver fibrosis in a large biopsyproven NAFLD cohort (n=427) in a follow-up study [13]. Four relevant eicosanoids were associated with fibrosis at baseline (11,12-dihydroxy-5Z,8Z,14Z,17Z-eicosatetraenoic acid [DIHETE], tetranor 12-hydroxyeicosatetraenoic acid [HETE], adrenic acid, and 14, 15-DIHETE). In addition, combination of changes in seven eicosanoids (5-HETE, 7,17-dihydroxy-(8Z,10,13,15E,19Z)-docosapentaenoic acid, adrenic acid, arachidonic acid, eicosapentaenoic acid, 16-hydroxy-docosahexaenoic acid, and 9-hydroxy-octadecadienoic acid) predicted improvement in fibrosis over 24-week period, suggesting plasma eicosanoids as potential biomarker of liver fibrosis.
Latest studies started to adopt more sophisticated approaches to derive metabolomics-based models to diagnose NASH and/or fibrosis. A proof-of-concept study by Perakakis et al. [14] conducted lipidomic, glycomic, and free fatty acid analyses from 49 controls and 31 biopsy-proven NAFLD patients (NAFLD, 15; NASH, 16). They elegantly derived a model consisting of 10 lipids which identified the presence of liver fibrosis with 98% accuracy [14]. Albeit not externally validated in large cohorts, the novel stepwise approach using supervised machine learning methods achieved more accurate results while reducing concerns on overfitting and overoptimism in the logistic regression analysis used in previous studies.
An intriguing study by Ogawa et al. [15] investigated metabolomic markers for hepatocellular ballooning, which is a characteristic histological feature of NASH [16]. Plasma phosphatidylcholine (aa-44:8) was significantly associated with the ballooning grade, which showed AUROC of 0.846 for the prediction in combination with type IV collagen 7S, choline, and lysophosphatidylethanolamine (e-18:2). These metabolomic markers seems promising, given that hepatocellular ballooning is a representative component for the diagnosis of NASH as well as monitoring of disease progression or resolution [17],
Lastly, a recent Korean study reported circulating alterations in lipidomic features which were distinctive between obese and nonobese NAFLD [18]. In this cross-sectional analysis of 361 biopsyproven NAFLD patients, visceral adiposity in nonobese NAFLD was significantly associated with saturated sphingomyelin (SM) species (SM d38:0), unlike in obese NAFLD. In addition, combinations of lipid metabolites showed good discriminative performance for nonobese (AUROC for NAFLD/NASH, 0.916/0.813) and obese (AUROC for NAFLD/NASH, 0.967/0.812) subjects, which included triacylglycerol (TAG) 46:1, TAG 48:1, TAG 50:1, SM d32:0, SM d38:0 for the nonobese group, and diacylglycerol (DAG) 34:1, DAG 40:7, DAG 40:8, TAG 46:1, TAG 48:1, TAG 50:2, SM d36:0 for the obese group, respectively. Although the cross-sectional design was unable to provide explicit information on the causal relationship, these lipid metabolites were deemed promising as novel biomarkers for nonobese NAFLD.
PLASMA AMINO ACID ALTERATIONS AND NAFLD SEVERITY
Protein and amino acid metabolism in the liver can affect glutathione synthesis, oxidative stress, insulin resistance, and inflammation. Alterations in circulating amino acids can be discovered frequently in patients with NAFLD, including increases in branchedchain amino acids (BCAAs; leucine, isoleucine, valine) and aromatic amino acids (AAAs; tryptophan, tyrosine, phenylalanine) and decrease in amino acids related to glutathione synthesis (glutamine, glycine, serine) [19-21]. Furthermore, plasma levels of BCAAs are increased in patients with obesity or T2DM [22,23], with higher levels of plasma BCAAs in men compared to women, suggesting sexual discrepancy in terms of susceptibilities to metabolic disorders [24].
Recent studies have reported the role of circulating amino acids as noninvasive biomarker for NAFLD severity. In Gaggini et al.’s study [20], hepatocellular ballooning and/or inflammation were associated with increased plasma BCAAs and AAAs, and fibrosis stages were discriminated using the levels of glutamate, serine and glycine. Grzych et al. [25] reported sex-dependent correlation between plasma BCAA levels and NAFLD severity, i.e., increased risk of NASH and fibrosis stage in women. Another study by Masarone et al. [26] investigated to differentiate a cohort of biopsy-proven NAFLD patients into NAFL, NASH, and NASH cirrhosis, using an untargeted plasma metabolomics profile and an ensemble machine learning. Relevant metabolites included increased glycocholic acid, taurocholic acid, phenylalanine, and BCAAs. Accuracy of the classification model for NAFL vs. NASH vs. NASH cirrhosis was 94.0%, with 94.1% sensitivity and 93.8% specificity.
NAFLD CHARACTERIZATION BASED ON BILE ACID ALTERATIONS
Role of bile acids in the pathogenesis of NAFLD has been unraveled in the last decade, particularly in metabolic homeostasis and insulin sensitivity. Dysregulation of bile acid metabolism is related to the development of dyslipidemia, insulin resistance, and hyperglycemia, all of which are common comorbidities of NAFLD [27]. Puri et al. [28] reported characteristic changes in plasma bile acid profile of NAFLD patients, showing significantly increased total primary bile acids and decreased secondary bile acids in NASH. Furthermore, this study also showed associations between bile acid alterations and histological severity of NASH, such as steatosis (taurocholate), lobular (glycocholate) and portal inflammation (taurolithocholate), hepatocyte ballooning (taurocholate), and fibrosis (total secondary to primary bile acid ratio and conjugated cholate). However, another study in obese subjects reported no correlation between plasma bile acid alterations and insulin resistance, without any correlation with NASH [29]. From a biopsy-proven cohort (n=102), Nimer et al. [30] reported plasma bile acids alteration, showing association of 7-keto-deoxycholic acid with advanced stages of fibrosis (odds ratio [OR], 4.2), NASH (OR, 24.5), and hepatocellular ballooning (OR, 18.7); 7-ketolithocholic acid with NASH (OR, 9.4) and ballooning (OR, 5.9). These results were contrary to another recent study by Caussy et al. [31], which reported inverse correlation between fibrosis and total free secondary bile acids. Collectively, it seems premature to conclude at the moment whether changes in bile acid metabolism are related to histological changes of NASH by playing relevant role in the pathogenesis or mere representation of metabolic comorbidities of NAFLD.
CONCLUSIONS AND FUTURE PERSPECTIVES
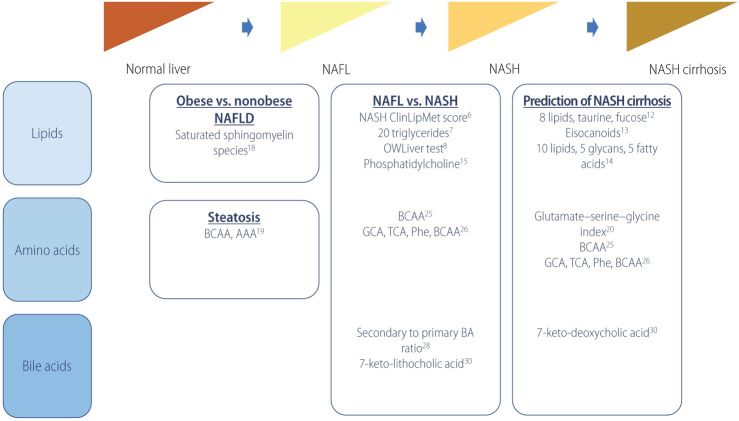
Noninvasive assessment of NAFLD phenotype or prediction of hepatic fibrosis has been extensively studied, which could reduce the necessity of biopsy for NAFLD patient at risk of NASH. Knowledge on the pathogenetic mechanism and development of high-throughput technologies in the last decade have accelerated studies on the metabolomics toward development of metabolitesbased noninvasive characterization of NAFLD. Table 1 and Figure 1 summarize metabolomics studies on the discrimination of NAFLD phenotypes as described above.
Table 1.
Recent metabolomics panels for the diagnosis of NAFL vs. NASH and prediction of fibrosis
| Study | Outcome | No. | Metabolites (panel) | Performance | |
|---|---|---|---|---|---|
| Lipid | |||||
| Zhou et al. [6] (2016) | Non-NASH vs. NASH | Estimation, n=223 (non-NASH, n=176; NASH, n=47) | NASH ClinLipMet Score (glutamate, isoleucine, glycine, lysophosphatidylcholine 16:0, phosphoethanolamine 40:6, AST, fasting insulin, PNPLA3 genotype) | AUROC, 0.866; sensitivity, 86%; specificity, 72% | |
| Validation, n=95 (non-NASH, n=71; NASH, n=24) | |||||
| Mayo et al. [7] (2018) | NAFL vs. NASH | Discovery, n=467 (control, n=90; NAFL, n=246; NASH, n=131) | 20 triglyceride species and BMI | AUROC, 0.95; sensitivity, 83%; specificity, 94% | |
| Validation, n=192 (control, n=7; NAFL, n=109; NASH, n=76) | |||||
| Barr et al. [8] (2012) | NAFL vs. NASH | Discovery (n=374) | BMI-dependent metabolic profile of 292 metabolites and 51 unidentified variables | AUROC, 0.84; sensitivity, 62%; specificity, 97% | |
| Validation (n=93) | |||||
| Bril et al. [9] (2018) | NAFL vs. NASH | 220 (no NAFLD, n=66; NAFL, n=39; NASH, n=115) | 20 triglyceride species and BMI | AUROC, 0.69 | |
| Caussy et al. [12] (2019) | F0–F2 vs. F3–F4 | Derivation, n=156 (F0–F2, n=133; F3–F4, n=23) | 8 lipids (5alpha-androstan-3beta monosulfate, pregnanediol-3-glucuronide, androsterone sulfate, epiandrosterone sulfate, palmitoleate, dehydroisoandrosterone sulfate, 5alphaandrostan-3beta disulfate, glycocholate), taurine, fucose | AUROC, 0.94; sensitivity, 90%; specificity, 79% | |
| Validation 1, n=142 (MRE <3.63 kPa, n=131; MRE ≥3.6 kPa, n=11) | |||||
| Validation 2 (n=59) | |||||
| Caussy et al. [13] (2020) | F0–F2 vs. F3–F4 | 427 (F0–2, n=229; F3–F4, n=197) | Baseline (11,12-DIHETE, tetranor 12-HETE, adrenic acid, and 14, 15-DIHETE), ≥1 stage improvement in fibrosis at 24-week follow-up (5-HETE, 7,17-DHDPA, adrenic acid, arachidonic acid, eicosapentaenoic acid, 16-HDOHE, and 9-HODE) | AUROC, 0.74 (follow-up) | |
| Perakakis et al. [14] (2019) | F0 vs. F1–F4 | 80 (control, n=49; NAFL, n=15; NASH, n=16) | 10 lipids, 5 glycans, 5 fatty acids | AUROC, 1.0; sensitivity, 97%; specificity, 99% | |
| Ogawa et al. [15] (2020) | Hepatocellular ballooning | 132 (non-ballooning, n=83; ballooning, n=49) | Type IV collagen 7S, choline, LPE (e-18:2) | AUROC, 0.846; sensitivity, 89.4%; specificity, 71.4% | |
| Amino acids | |||||
| Gaggini et al. [20] (2018) | F0–F2 vs. F3–F4 | 64 (control, n=20; nonobese NAFLD, n=29; obese NAFLD, n=15) | Glutamate-serine-glycine index | OR, 122 (P=0.004) | |
| Grzych et al. [25] (2020) | NAFL vs. NASH vs. NASH cirrhosis | Discovery, n=213 (control, n=69; NAFL, n=78; NASH, n=23; NASH cirrhosis, n=15; non-NASH cirrhosis, n=28) | Glycocholic acid, taurocholic acid, phenylalanine, branched-chain amino-acids | Accuracy, 94.0%; sensitivity, 94.1%; specificity, 93.8% | |
| Validation, n=94 (control, n=44; NAFL, n=34; NASH, n=10; NASH-cirrhosis, n=6) | |||||
| Bile acid | |||||
| Legry et al. [29] (2017) | Advanced fibrosis, NASH, hepatocellular ballooning | 152 (control, n=50; NAFLD, n=102) | Keto-deoxycholic acid, 7-ketolithocholic acid | Keto-deoxycholic acid with advanced fibrosis (OR, 4.2), NASH (OR, 24.5) and hepatocellular ballooning (OR, 18.7); 7-ketolithocholic acid with NASH (OR, 9.4) and ballooning (OR, 5.9) | |
F0–F4 represents the stage of hepatic fibrosis on a scale from 0 to 4.
NAFL, nonalcoholic fatty liver; NASH, nonalcoholic steatohepatitis; AST, aspartate aminotransferase; PNPLA3, patatin-like phospholipase domaincontaining-3; AUROC, area under the receiver operating characteristic; BMI, body mass index; NAFLD, nonalcoholic fatty liver disease; MRE, magnetic resonance elastography; DIHETE, dihydroxy-5Z,8Z,14Z,17Z-eicosatetraenoic acid; HETE, hydroxyeicosatetraenoic acid; DHDPA, dihydroxy-(8Z,10,13,15E,19Z)-docosapentaenoic acid; HDOHE, hydroxy-docosahexaenoic acid; HODE, hydroxy-octadecadienoic acid; LPE, lysophosphatidylethanolamine; OR, odds ratio.
Figure 1.
Metabolomics and prediction of NAFLD phenotypes. NAFL, nonalcoholic fatty liver; NASH, nonalcoholic steatohepatitis; NAFLD, nonalcoholic fatty liver disease; BCAA, branched-chain amino acid; AAA, aromatic amino acid; GCA, glycocholic acid; TCA, taurocholic acid; Phe, phenylalanine; BA, bile acid.
To date, despite many promising biomarker candidates from numerous recent studies, there is no widely acknowledged metabolomics marker for NAFLD phenotype or severity. Possible explanations for the paucity of accurate and precise marker include the followings. First, NAFLD is a multisystem disease involving complex interplay of genetic predisposition, lifestyle risk factors such as lack of physical activity and/or unhealthy diet habit, presence of comorbid metabolic diseases, and gut microbiota [32]. All these components determine the phenotypes and severity of NAFLD, while affecting metabolites at the same time. Thus, adjustment for those confounding factors and consideration of population heterogeneity are required in the design of NAFLD metabolomics study. Second, most studies on NAFLD biomarkers were crosssectional design, from limited size of study population. Prospective validation in larger-size independent cohorts with diverse characteristics is warranted for potential biomarkers with promising performance. Finally, comprehensive mechanistic studies are warranted to disentangle the roles of metabolites in NASH pathogenesis and their significance as novel pharmacological targets. In addition, associations between genetic polymorphisms, such as PNPLA3, and metabolomic findings need to be explored in future studies.
In conclusion, metabolomics research has provided important insights in the pathophysiology of NAFLD. Integration of metabolomics data and clinical information will hopefully lead to discovery of individual molecular signature in patients with NAFLD and thereby identification of patients at risk of development and/or disease progression of NAFLD between patient subgroups [33]. However, personalized medicine mandates rigorous validation and replication before implementation of metabolomics-derived biomarkers in the management of patients with NAFLD, on top of detailed characterization of disease phenotypes and molecular features [34]. Collectively, identification of the phenotype-specific metabolomics biomarkers is anticipated to contribute toward personalized medicine in the field of NAFLD. For this purpose, greater understanding of underlying mechanism in the evolution of NAFL to NASH and unraveling metabolite biomarkers involved in NASH pathogenesis might lead to the discovery of novel diagnostic as well as therapeutic options.
Acknowledgments
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (Grant no. 2018R1A5A2025286), the Research Supporting Program of the Korean Association for the Study of the Liver (Grant no. KASL2018-01) and the Technology Innovation Program (or Industrial Strategic Technology Development Program-Bioindustry Strategic Technology Development Program) (Grant no. 2013712) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
Abbreviations
- AAA
aromatic amino acid
- AST
aspartate aminotransferase
- AUROC
area under the receiver operating characteristic
- BCAA
branched-chain amino acid
- DAG
diacylglycerol
- DIHETE
dihydroxy-5Z
- HETE
hydroxyeicosatetraenoic acid
- MS
mass spectrometry
- NAFL
nonalcoholic fatty liver
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- OR
odds ratio
- PNPLA3
patatin-like phospholipase domain-containing-3
- SM
sphingomyelin
- T2DM
type 2 diabetes mellitus
- TAG
triacylglycerol
Footnotes
Conflicts of Interest: The author has no conflicts to disclose.
REFERENCES
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 3.Brunt EM, Kleiner DE, Carpenter DH, Rinella M, Harrison SA, Loomba R, et al. NAFLD: reporting histologic findings in clinical practice. Hepatology. 2021;73:2028–2038. doi: 10.1002/hep.31599. [DOI] [PubMed] [Google Scholar]
- 4.Verhaegh P, Bavalia R, Winkens B, Masclee A, Jonkers D, Koek G. Noninvasive tests do not accurately differentiate nonalcoholic steatohepatitis from simple steatosis: a systematic review and metaanalysis. Clin Gastroenterol Hepatol. 2018;16:837–861. doi: 10.1016/j.cgh.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 5.Dumas ME, Kinross J, Nicholson JK. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology. 2014;146:46–62. doi: 10.1053/j.gastro.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Oresic M, Leivonen M, Gopalacharyulu P, Hyysalo J, Arola J, et al. Noninvasive detection of nonalcoholic steatohepatitis using clinical markers and circulating levels of lipids and metabolites. Clin Gastroenterol Hepatol. 2016;14:1463–1472.e6. doi: 10.1016/j.cgh.2016.05.046. [DOI] [PubMed] [Google Scholar]
- 7.Mayo R, Caballería J, Martínez-Arranz I, Banales JM, Arias M, Mincholé I, et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: results from discovery and validation cohorts. Hepatol Commun. 2018;2:807–820. doi: 10.1002/hep4.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barr J, Caballería J, Martínez-Arranz I, Domínguez-Díez A, Alonso C, Muntané J, et al. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J Proteome Res. 2012;11:2521–2532. doi: 10.1021/pr201223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bril F, Millán L, Kalavalapalli S, McPhaul MJ, Caulfield MP, MartinezArranz I, et al. Use of a metabolomic approach to non-invasively diagnose non-alcoholic fatty liver disease in patients with type 2 diabetes mellitus. Diabetes Obes Metab. 2018;20:1702–1709. doi: 10.1111/dom.13285. [DOI] [PubMed] [Google Scholar]
- 10.Ekstedt M, Hagström H, Nasr P, Fredrikson M, Stål P, Kechagias S, Hultcrantz R. Fibrosis stage is the strongest predictor for diseasespecific mortality in NAFLD after up to 33 years of follow-up. Hepatology. 2015;61:1547–1554. doi: 10.1002/hep.27368. [DOI] [PubMed] [Google Scholar]
- 11.Dulai PS, Singh S, Patel J, Soni M, Prokop LJ, Younossi Z, et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta-analysis. Hepatology. 2017;65:1557–1565. doi: 10.1002/hep.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caussy C, Ajmera VH, Puri P, Hsu CL, Bassirian S, Mgdsyan M, et al. Serum metabolites detect the presence of advanced fibrosis in derivation and validation cohorts of patients with non-alcoholic fatty liver disease. Gut. 2019;68:1884–1892. doi: 10.1136/gutjnl-2018-317584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caussy C, Chuang JC, Billin A, Hu T, Wang Y, Subramanian GM, et al. Plasma eicosanoids as noninvasive biomarkers of liver fibrosis in patients with nonalcoholic steatohepatitis. Therap Adv Gastroenterol. 2020;13:1756284820923904. doi: 10.1177/1756284820923904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perakakis N, Polyzos SA, Yazdani A, Sala-Vila A, Kountouras J, Anastasilakis AD, et al. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: a proof of concept study. Metabolism. 2019;101:154005. doi: 10.1016/j.metabol.2019.154005. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa Y, Kobayashi T, Honda Y, Kessoku T, Tomeno W, Imajo K, et al. Metabolomic/lipidomic-based analysis of plasma to diagnose hepatocellular ballooning in patients with non-alcoholic fatty liver disease: a multicenter study. Hepatol Res. 2020;50:955–965. doi: 10.1111/hepr.13528. [DOI] [PubMed] [Google Scholar]
- 16.Kleiner DE, Makhlouf HR. Histology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis in adults and children. Clin Liver Dis. 2016;20:293–312. doi: 10.1016/j.cld.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratziu V. A critical review of endpoints for non-cirrhotic NASH therapeutic trials. J Hepatol. 2018;68:353–361. doi: 10.1016/j.jhep.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Jung Y, Lee MK, Puri P, Koo BK, Joo SK, Jang SY, et al. Circulating lipidomic alterations in obese and non-obese subjects with nonalcoholic fatty liver disease. Aliment Pharmacol Ther. 2020;52:1603–1614. doi: 10.1111/apt.16066. [DOI] [PubMed] [Google Scholar]
- 19.Yamakado M, Tanaka T, Nagao K, Imaizumi A, Komatsu M, Daimon T, et al. Plasma amino acid profile associated with fatty liver disease and co-occurrence of metabolic risk factors. Sci Rep. 2017;7:14485. doi: 10.1038/s41598-017-14974-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaggini M, Carli F, Rosso C, Buzzigoli E, Marietti M, Della Latta V, et al. Altered amino acid concentrations in NAFLD: impact of obesity and insulin resistance. Hepatology. 2018;67:145–158. doi: 10.1002/hep.29465. [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa T, Iino C, Endo T, Mikami K, Kimura M, Sawada N, et al. Changed amino acids in NAFLD and liver fibrosis: a large crosssectional study without influence of insulin resistance. Nutrients. 2020;12:1450. doi: 10.3390/nu12051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laferrère B, Reilly D, Arias S, Swerdlow N, Gorroochurn P, Bawa B, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Sci Transl Med. 2011;3:80re2. doi: 10.1126/scitranslmed.3002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krumsiek J, Mittelstrass K, Do KT, Stückler F, Ried J, Adamski J, et al. Gender-specific pathway differences in the human serum metabolome. Metabolomics. 2015;11:1815–1833. doi: 10.1007/s11306-015-0829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grzych G, Vonghia L, Bout MA, Weyler J, Verrijken A, Dirinck E, et al. Plasma BCAA changes in patients with NAFLD are sex dependent. J Clin Endocrinol Metab. 2020;105:2311–2321. doi: 10.1210/clinem/dgaa175. [DOI] [PubMed] [Google Scholar]
- 26.Masarone M, Troisi J, Aglitti A, Torre P, Colucci A, Dallio M, et al. Untargeted metabolomics as a diagnostic tool in NAFLD: discrimination of steatosis, steatohepatitis and cirrhosis. Metabolomics. 2021;17:12. doi: 10.1007/s11306-020-01756-1. [DOI] [PubMed] [Google Scholar]
- 27.Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes. 2013;62:4184–4191. doi: 10.2337/db13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puri P, Daita K, Joyce A, Mirshahi F, Santhekadur PK, Cazanave S, et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology. 2018;67:534–548. doi: 10.1002/hep.29359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legry V, Francque S, Haas JT, Verrijken A, Caron S, Chávez-Talavera O, et al. Bile acid alterations are associated with insulin resistance, but not with NASH, in obese subjects. J Clin Endocrinol Metab. 2017;102:3783–3794. doi: 10.1210/jc.2017-01397. [DOI] [PubMed] [Google Scholar]
- 30.Nimer N, Choucair I, Wang Z, Nemet I, Li L, Gukasyan J, et al. Bile acids profile, histopathological indices and genetic variants for non-alcoholic fatty liver disease progression. Metabolism. 2021;116:154457. doi: 10.1016/j.metabol.2020.154457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caussy C, Hsu C, Singh S, Bassirian S, Kolar J, Faulkner C, et al. Serum bile acid patterns are associated with the presence of NAFLD in twins, and dose-dependent changes with increase in fibrosis stage in patients with biopsy-proven NAFLD. Aliment Pharmacol Ther. 2019;49:183–193. doi: 10.1111/apt.15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powell EE, Wong VW, Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397:2212–2224. doi: 10.1016/S0140-6736(20)32511-3. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen J. Systems biology of metabolism: a driver for developing personalized and precision medicine. Cell Metab. 2017;25:572–579. doi: 10.1016/j.cmet.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Bjornson E, Borén J, Mardinoglu A. Personalized cardiovascular disease prediction and treatment-a review of existing strategies and novel systems medicine tools. Front Physiol. 2016;7:2. doi: 10.3389/fphys.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]