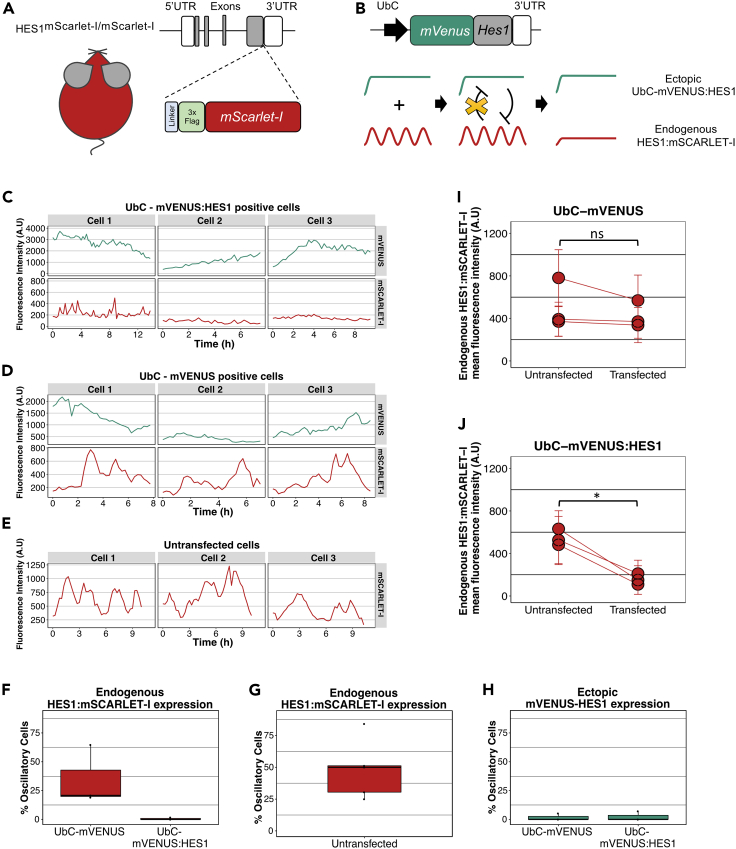
Figure 3.
Endogenous HES1 protein does not oscillate in the presence of an ectopic HES1 persistent input
(A) Schematic structure of the Hes1 locus in the HES1mScarlet-I/mScarlet-I transgenic mice. A DNA sequence encoding for a linker protein, a 3xFlag epitope and the mSACRLET-I protein has been inserted downstream of the last Hes1 exon and before the 3′UTR.
(B) Schematic showing the structure of UbC-mVENUS:HES1 reporter and the hypothesis for altering the endogenous HES1:mSCARLET-I oscillatory expression and level. mVenus has been fused to Hes1 cDNA followed by the Hes1 3′UTR and it is expressed under the constitutive UbC promoter. Persistent HES1 expression is expected to inhibit the endogenous HES1 level and override the endogenous HES1 oscillations.
(C–E) Example cell traces showing mVENUS and mSCARLET-I fluorescence expression over time in the same cell where mVENUS represents expression of either UbC-mVENUS:HES1 (C) or the control UbC-mVENUS (D) and mSCARLET-I represents endogenous HES1 expression. In untransfected cells only the mSCARLET-I endogenous HES1 expression has been recorded (E) (A.U. = arbitrary units). Oscillatory period of mSCARLET-I expression in panel (D) Cell 1 = 2.4h, Cell 2 = 2.4h, Cell 3 = 3h and in panel (E) Cell 1 =3.2h, Cell 2 =3.1h, Cell 3 = 5.3h.
(F and G) Box plots showing the percentage of cells that have oscillatory endogenous HES1:mSCARLET-I expression in the presence of ectopic UbC-mVENUS or UbC-mVENUS:HES1 (F) or in untransfected cells (G) (dots represent means of biological replicates, black horizontal lines represent median, n = 3 biological experiments).
(H) Box plot showing the percentage of cells where UbC-mVENUS or UbC-mVENUS:HES1 exhibit oscillatory expression (dots represent means of biological replicates, black horizontal lines represent median, n = 3 biological experiments).
(I and J) Graphs showing the endogenous HES1:mSCARLET-I expression level in untransfected vs transfected cells with UbC-mVENUS (I) or UbC-mVENUS:HES1 reporter (J) per experiment (circles represent mean values and error bars represent standard deviation, A.U. = arbitrary units). Number of cells analyzed for (I) untransfected = 191, transfected = 135, n = 3 biological experiments, two-tailed paired ttest, ns = not significant. Number of cells analyzed for (J) untransfected = 222, transfected = 88, n = 3 biological experiments, two-tailed paired ttest, ∗p = 0.015.