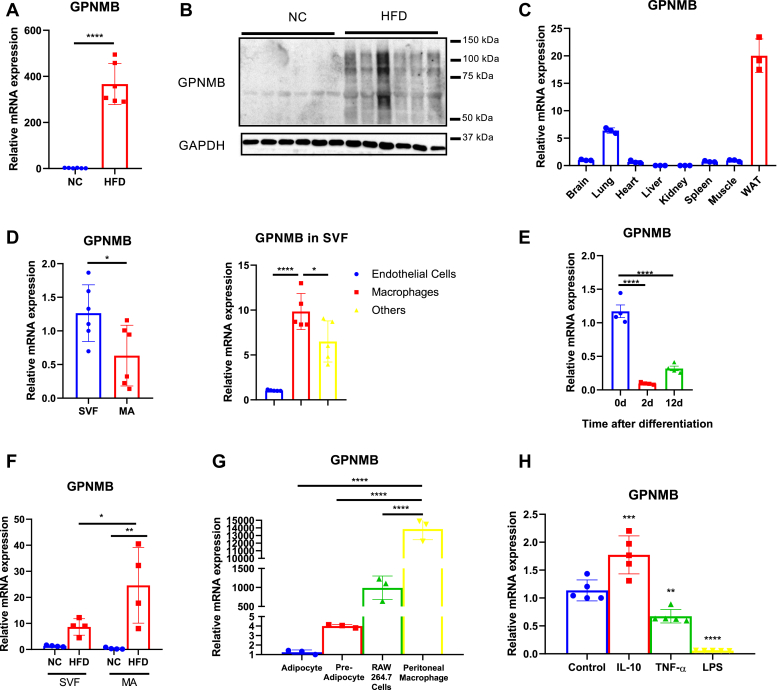
Figure 1.
GPNMB expression is robustly increased in the WAT of obese mice.A, quantitative PCR for GPNMB in the WAT isolated from either lean mice fed normal chow (NC) or obese mice fed an high-fat diet (HFD) for 12 weeks (n = 6 each). B, immunoblotting for GPNMB in the WAT isolated from lean mice fed a NC or obese mice fed an HFD (n = 6 each). C, quantitative PCR for GPNMB in various tissues of lean mice (n = 3 each). D, quantitative PCR for GPNMB in stromal vascular fraction (SVF) and mature adipocyte (MA) of the WAT isolated from lean mice (n = 6 each; left). Cells in SVF were separated into endothelial cells, macrophages, and others by using magnetic-activated cell sorting. GPNMB expressions in these cells were analyzed (n = 5 each; right). E, quantitative PCR for GPNMB in 3T3-L1 preadipocytes at the indicated time after induction of adipocyte differentiation (n = 4 each). F, quantitative PCR for GPNMB in SVF and MA of the WAT isolated from lean mice fed NC or obese mice fed an HFD (n = 4 each). G, quantitative PCR for GPNMB in 3T3L1-derived adipocytes, 3T3L1 preadipocytes, RAW264.7 macrophages, and mouse resident peritoneal macrophages (n = 3 each). H, mouse resident peritoneal macrophages were treated either vehicle (control), IL-10 (10 ng/ml), TNF-α (10 ng/ml) or LPS (10 ng/ml) for 24 h, and then GPNMB expression was analyzed (n = 5 each). Data represent mean ± SDM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001. Two-tailed Student’s t test was used for the analysis of the differences between the groups (A and D left), while one-way ANOVA with Tukey’s post hoc test for multiple comparisons was used for the analysis of the differences between each group (D right, E–H).