FIGURE 1.
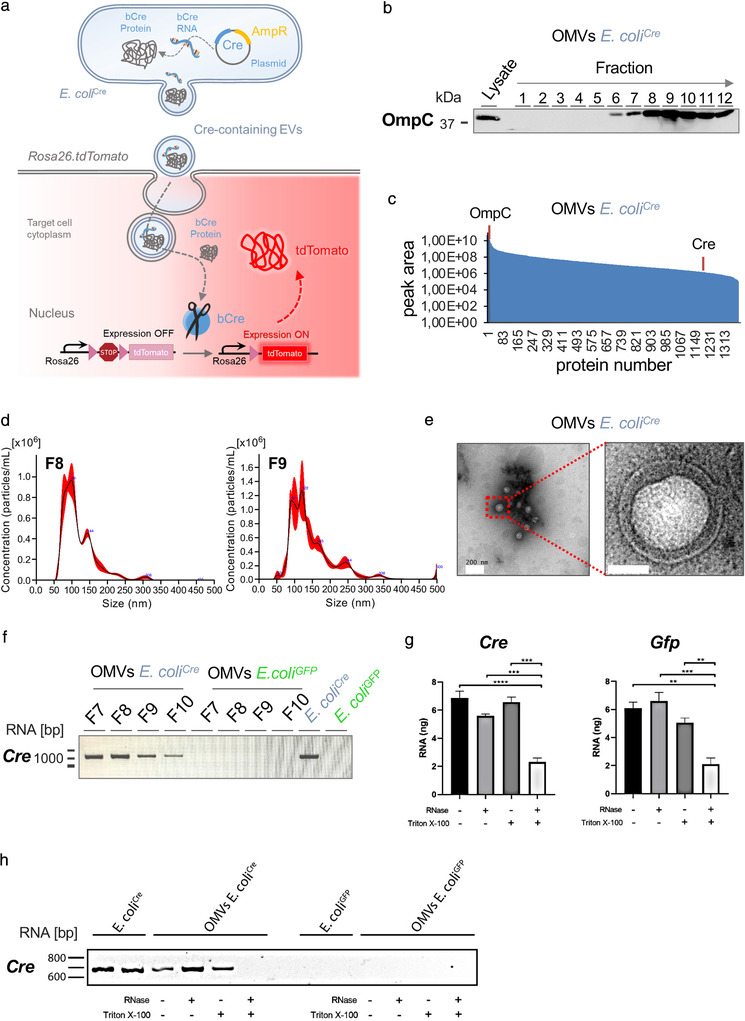
Characterization of OMVs. (a) Graphical illustration of bacterial OMVs transferring their molecular cargo to mammalian host cells. E. coli Cre‐derived OMVs containing Cre RNA or protein are taken up by host cells of Rosa26.tdTomato reporter mice, leading to Cre‐mediated excision of the loxP‐flanked STOP cassette, thereby inducing expression of robust tdTomato fluorescence (red). (b) Western blot analysis using 37,5 μl of each fraction derived from iodixanol density gradient purification as well as bacterial lysates (5 μg) of E. coli Cre. The membrane was probed with an antibody against OmpC (37 kDa). (c) Dynamic range of OMV contained proteins analyzed by mass spectrometry, relative quantity of OmpC and Cre are indicated by orange lines. (d) Representative NTA analysis of fraction 8 and 9 of iodixanol density gradient purified E. coli Cre OMVs. (e) Transmission electron microscopic image of OMV preparation isolated from E. coli Cre culture supernatant showing membranous particles confirming the presence of small OMVs. Scale Bar: 200 nm and 50 nm. (f) RT‐PCR analysis of full length Cre RNA from fractions F7 to F10 of iodixanol density gradient purified OMVs and whole bacterial lysates from E. coli Cre and E. coli GFP. (g) Isolated OMVs (OMVs E. coli Cre and OMVs E. coli GFP) were left unchallenged or treated with RNAse or Triton X‐100 or a combination of the two. RNA concentration was measured by Quant‐iT RiboGreen RNA Assay. (h) RT‐PCR analysis of Cre RNA from whole bacterial lysates and iodixanol density gradient purified OMVs (from E. coli Cre and E. coli GFP) left unchallenged or treated with RNAse, Triton X‐100 or a combination of both.
