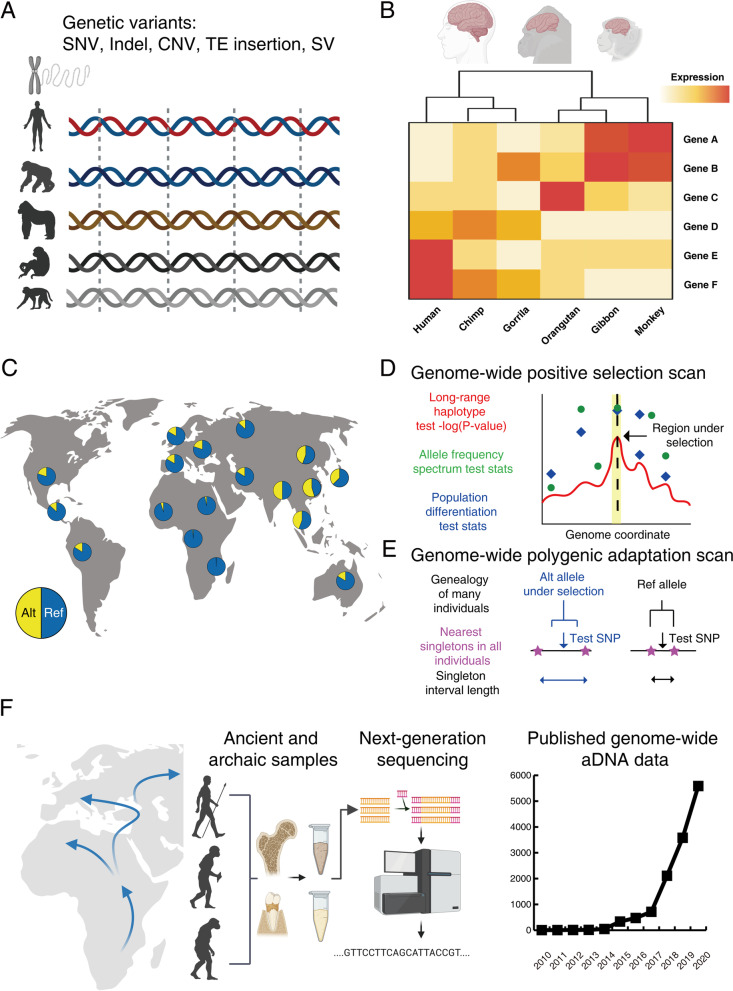
Fig. 1.
Computational approaches to study the genetic basis of human brain evolution. (A) Comparative studies of human and closely related non-human primates (NHPs). Cross-species genomic comparison reveals various types of genetic changes ranging from single nucleotide variants (SNVs) to megabase-scale copy number and structural variants (CNVs and SVs) in orthologous genes and non-coding regions. (B) Cross-species transcriptomic analysis identifies spatial and temporal changes in gene expression and RNA splicing during brain development and evolution. (C) Population genetic analysis of diverse modern human populations. Differences in allele frequencies of a polymorphic variant (yellow in the pie charts) across different populations may indicate natural selection. (D) Genome-wide positive selection scans, including the long-range haplotype test, the allele frequency spectrum test, and the population differentiation test predict genomic regions under positive selection. (E) The Singleton-Density-Score method identifies genomic regions under polygenic adaptation by detecting alternative alleles that show unusually short terminal branch lengths due to long intervals between singleton test SNPs in the genealogy tree, compared to the lengths of the corresponding reference alleles. (F) Leveraging ancient human genomes to identify genetic variants with allele frequency changes over time. The number of ancient/archaic human samples (WGS and SNP arrays) are shown according to publication years