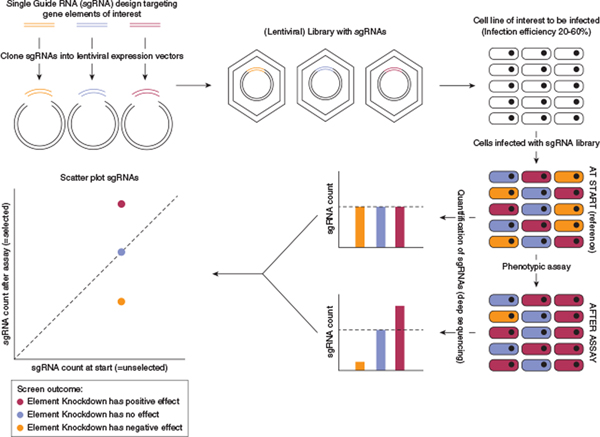
Figure 1. Overview of the key steps during a typical CRISPR-ko screen.
CRISPR sgRNA libraries can be ordered commercially, obtained through public repositories (e.g., Addgene), or custom-designed. For custom CRISPR libraries, pooled sgRNAs can be ordered as a DNA oligonucleotide pool. After PCR amplification, the library is cloned into a delivery vector (e.g., a lentiviral vector). Following packaging into lentiviral particles, the CRISPR library is infected into target cells at an infection efficiency of 20–60% to maximize the percentage of cells transduced with a single sgRNA. After infection, cells are selected with an antibiotic to deplete noninfected cells. A subset of cells is then collected as reference (at start, reference and/or unselected). An assay is then applied to select for cells displaying a desired phenotype, and cells are harvested at the endpoint and optionally at intermediate timepoints. Genomic DNA of both reference and endpoint cells is isolated and primers flanking the sgRNAs are deep sequenced from bulk DNA to measure the abundance of each sgRNA (sgRNA count) at each timepoint. sgRNA abundances can be visualized by plotting the sgRNA counts pre- and post-screen. Negative control sgRNAs (without biological targets) should appear around the dotted line, representing no change. CRISPR-ko, CRISPR knockout; sgRNA, single guide RNA.