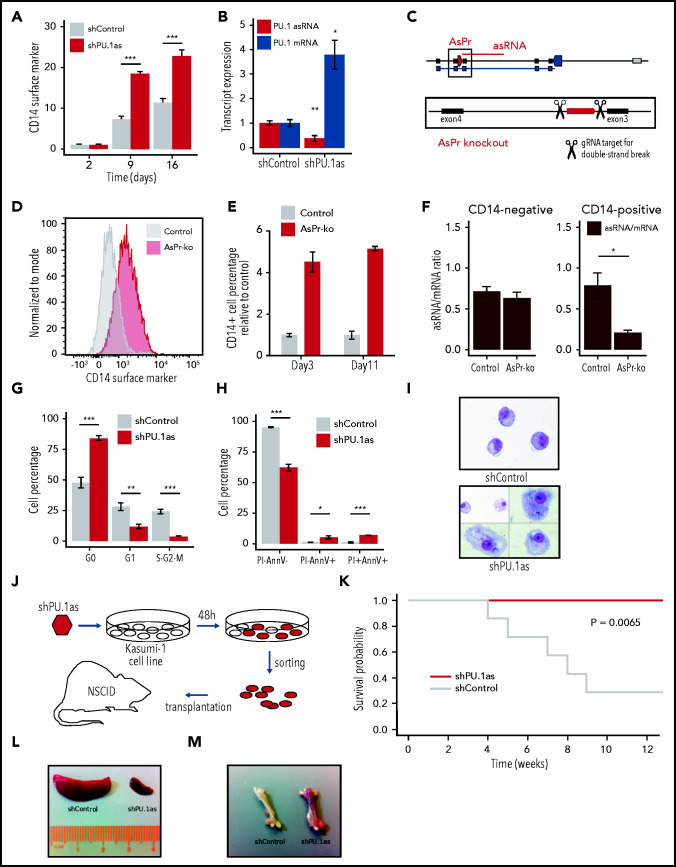
Figure 4.
PU.1 antisense drives leukemia in RUNX1-ETO cells. (A) Fluorescence-activated cell sorting (FACS) analysis for CD14 surface marker expression and cell viability kinetics after shRNA-mediated knockdown shPU.1as in Kasumi-1 (n = 4). (B) PU.1 mRNA and asRNA expression in Kasumi-1 after shPU.1as in Kasumi-1 (n = 4). Also shown is the antisense transcription start site. (C) Schematic representation of the PU.1 locus with the AsPr (red arrow box) regulating asRNA (red line) and the double-strand break locations for CRISPR/Cas9-mediated AsPr knockout. (D) FACS analysis for CD14 surface marker expression after AsPr knockout (AsPr-ko) in Kasumi-1 (merged n = 4). (E) Kinetics of CD14 surface marker expression after AsPr-ko in Kasumi-1 (n = 4). (F) PU.1 asRNA/mRNA ratio (brown) after AsPr-ko in CD14– and CD14+ Kasumi-1 cells (n = 4). Assays for cell cycle stages after shPU.1as (G) and cell viability (H) (n = 4) in Kasumi-1 cells (shControl, n = 5; shPU.1as, n = 6). (I) May Grünwald/Giemsa cytospins for morphology analysis of Kasumi-1 cells. (J) Experimental workflow of shRNA knockdown of PU.1 antisense RNA (shPU.1as) in Kasumi-1 followed by FACS and xenografting into immunodeficient (NOD/SCID) mice. (K) Survival probability of NOD-SCID mice xenografted with Kasumi-1 cells after shPU.1as vs shControl (n = 7). Leukemic cells infiltration in spleen (L) and bone (M) samples from mice transplanted with Kasumi-1 cells. Data are represented as mean ± standard error of the mean. *P < .05, **P < .01, ***P < .001, Student t test. AnnV, Annexin V; PI, propidium iodide.