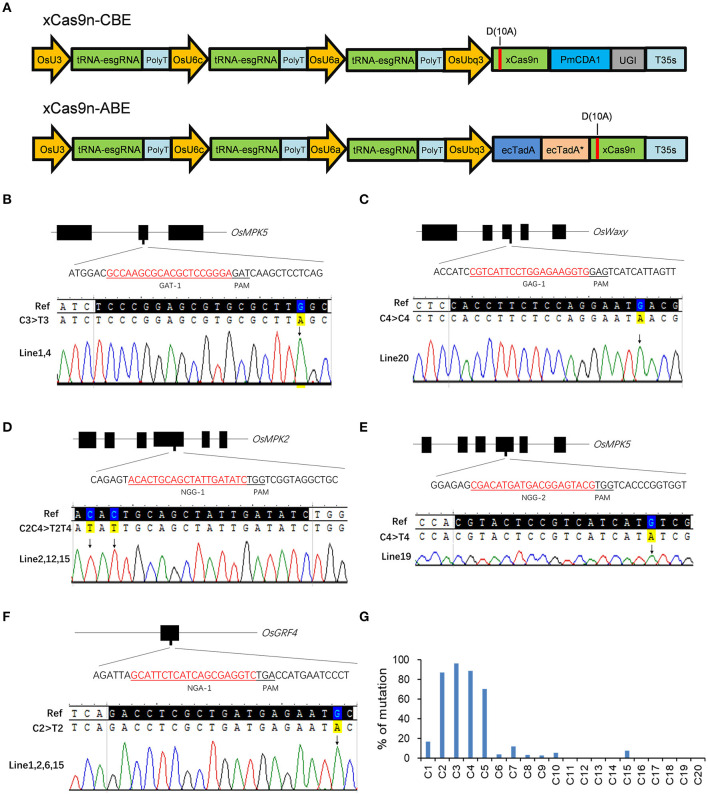
Figure 3.
Base editing by xCas9 in rice T0 plants. (A) Schematic view of xCas9n-CBE and xCas9n-ABE. (B–F) Sequencing chromatograms of all homozygous mutant lines in which xCas9n-CBE modified the GAT-1 (B), GAG-1 (C), NGG-1 (D), NGG-2 (E), and NGA-1 (F) target sites. The mutated bases are marked by arrows. (G) Ratio of T0 plants mutated at a certain position to all genome-edited T0 plants containing C at that position. The x-axis presents the position of C from the 5′ end of the target.