Abstract
Inherited bone marrow failure syndromes (IBMFS) are monogenetic disorders that result in a reduction of mature blood cell formation and predisposition to leukemia. In children with myeloid leukemia the gene most often mutated is Gata binding protein 2 (GATA2) and 80% of patients with GATA2 mutations develop myeloid malignancy before the age of forty. Although GATA2 is established as one of the key regulators of embryonic and adult hematopoiesis, the mechanisms behind the leukemia predisposition in GATA2 haploinsufficiencies is ambiguous. The only curative treatment option currently available is allogeneic hematopoietic stem cell transplantation (allo-SCT). However, allo-SCT can only be applied at a relatively late stage of the disease as its applicability is compromised by treatment related morbidity and mortality (TRM). Alternatively, autologous hematopoietic stem cell transplantation (auto-SCT), which is associated with significantly less TRM, might become a treatment option if repaired hematopoietic stem cells would be available. Here we discuss the recent literature on leukemia predisposition syndromes caused by GATA2 mutations, current knowledge on the function of GATA2 in the hematopoietic system and advantages and pitfalls of potential treatment options provided by genome editing.
Keywords: GATA2, Inherited bone marrow failure syndrome, MDS, AML, GATA2 haploinsufficiency syndrome, genome editing, HSCs, autologous HSC transplantation
Introduction
IBMFS are a heterogeneous cluster of disorders manifested by an ineffective blood production and concurrent cytopenias that eventually result in a hypoplastic bone marrow (BM). These syndromes constitute an increased propensity to develop hematological malignancies such as myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) (Dokal and Vulliamy, 2010; Wilson et al., 2014; Cook, 2018). Mutations in GATA2 are the most common genetic defects in pediatric MDS (Spinner et al., 2014). GATA2 is one of the master regulators of blood production and patients that carry a mutation in one of the two alleles of GATA2 often manifest with immunodeficiency syndromes and increased lifetime risk for MDS/AML (Wlodarski et al., 2016; Donadieu et al., 2018; McReynolds et al., 2018). Once malignant transformation becomes overt, survival rates are below 50% (Spinner et al., 2014). Due to the inherited mutation, allo-SCT is the only curative treatment option for these patients (Simonis et al., 2018; van Lier et al., 2020). Unfortunately, the use of allo-SCT is compromised by TRM and not applicable for patients who have not progressed to leukemia yet. Uncovering the modus operandi of GATA2 and other (epi)genetic factors in the complex network of blood regulation is essential to design non-invasive and preventive treatment options for IBMFS patients.
Genome editing strategies, especially the implementation of clustered regularly interspaced short palindromic repeat/associated protein 9 (CRISPR/Cas9) nuclease platforms, improve rapidly and progress toward efficient therapies for several genetic diseases (Cong et al., 2013; Mali et al., 2013; Anzalone et al., 2019). In this review, we will summarize clinical symptoms of GATA2 haploinsufficiency patients and results from Gata2 experimental models to inspect the function of GATA2 in leukemogenesis. Our aim is to explore the potential and pitfalls of genome editing methods to treat GATA2 deficiency syndromes in the light of current technologies.
The Transcription Factor GATA2
GATA2 is a zinc finger transcription factor that contains 2 first exons; a hematopoietic and neuronal cell specific distal first exon and a proximal first exon that is utilized ubiquitously. These two transcript variants encode the same protein (Minegishi et al., 1998; Pan et al., 2000). GATA2 binds a highly conserved (A/T)GATA(A/G) DNA sequence and other protein partners through two multifunctional zinc finger (ZF) domains; ZF1 and ZF2 that are encoded by exon 4 and exon 5, respectively (Evans and Felsenfeld, 1989; Alfayez et al., 2019). Two GATA2 protein isoforms can be formed, one lacking exon 5 and consequently lacking the ZF2 domain (Vicente et al., 2012) (Figure 1). To date, the functional consequence of this remains unclear.
Figure 1.
GATA2 locus organization and overview of mutation types found in GATA2 haploinsufficiency patients.
Germline GATA2 Mutations
In 2011, four different studies described germline heterozygous GATA2 mutations in a total of 44 patients with various syndromes; monocytopenia and mycobacterial infection (MonoMAC) syndrome (Hsu et al., 2011), monocyte, B cell, NK cell and dendritic cell deficiencies (DCML) (Dickinson et al., 2011), Emberger Syndrome, which is characterized by primary lymphedema with a predisposition to AML (Ostergaard et al., 2011) and familial MDS/AML predisposition (Hahn et al., 2011). Distinct clinical perspectives discerned in these studies coalesce under the theme of the loss of one allele of GATA2 resulting in the GATA2 haploinsufficiency syndrome, which can present with immunodeficiency, lymphedema and 80% predisposition to develop MDS/AML.
Taken together, 60% of patients present with a truncating mutation in GATA2 before the ZF2 domain and 30% of patients present with a non-synonymous mutation in ZF2. However, some patients develop MonoMAC syndrome without mutations in the coding region of GATA2 but have reduced GATA2 expression levels (Hsu et al., 2013). These patients harbor mutations in the intronic region, specifically in intron 4. Mutations in this region abrogate the function of a conserved +9.5 cis-element, that regulates GATA2 transcription levels resulting in GATA2 haploinsufficiency (Hsu et al., 2013) and intron 4 mutations represent 10% of all GATA2 haploinsufficiency cases (Wlodarski et al., 2017) (Figure 1). GATA2 mutations are also present in a subset of patients with chronic neutropenia and aplastic anemia (AA) (Townsley et al., 2012; Pasquet et al., 2013). However, BM of AA patients with GATA2 mutations encompasses noticeably different types of altered hematopoietic populations than idiopathic AA patients, such as the complete loss of lymphoid progenitors and atypical megakaryocytes (Ganapathi et al., 2015).
Both familial and sporadic mutations in the coding and cis-regulatory regions of GATA2 are found and are the underlying cause in 15% of advanced and 7% of all pediatric MDS cases (Wlodarski et al., 2016). Most of these mutations can be found in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar). Currently, despite the improving definition of the phenotypic characteristics of GATA2 deficiency syndromes and high penetrance of myeloid malignancy, the mutational background and phenotypic outcome observed in these patients do not correlate, suggesting that additional events are important for disease progression (Collin et al., 2015; Wlodarski et al., 2016). Evidence for this is found in a cohort of pediatric MDS-GATA2 patients that acquired additional somatic mutations in ASXL1, RUNX1, SETBP1, IKZF1, and CRLF2 genes, which resulted in an increased progression to AML. Furthermore, 72% of adolescents with MDS and monosomy 7 had an underlying GATA2 mutation (Wlodarski et al., 2016; Fisher et al., 2017; Yoshida et al., 2020).
Somatic GATA2 Mutations
Although truncating germline GATA2 mutations occur most often, a few somatic mutations are reported that phenocopy germline loss-of-function mutations (Sekhar et al., 2018; Alfayez et al., 2019). These cause a relatively milder form of the immunodeficiency phenotype observed in germline mutant GATA2 patients, along with a common presentation of AML, atypical chronic myeloid leukemia and in some cases acute erythroid leukemia (Ping et al., 2017; Sekhar et al., 2018; Alfayez et al., 2019).
Somatic GATA2 mutations are found both in ZF1 and ZF2 and all patients with somatic GATA2 mutations harbor mutations in other genes, predominantly CEBPA with an incidence of 18–21% (Fasan et al., 2013; Hou et al., 2015; Theis et al., 2016). In one cohort of AML patients, ZF1 but not ZF2 mutations in GATA2 closely associate with biallelic CEBPA mutations (Tien et al., 2018). This implies that ZF1 is crucial for GATA2 function in disease progression in combination with CEBPA mutations.
The Function of the Transcription Factor GATA2 in Mammalian Hematopoiesis
The Function of GATA2 in Embryonic Hematopoiesis
In mouse, homozygous deletion of Gata2 results in 67% lethality at embryonic day (E) 10.5 and none survive beyond E11.5, due to severe anemia. Chimeras of WT and Gata2−/− embryonic stem (ES) cells show that Gata2-null cells cannot contribute to hematopoiesis in adult blood, fetal liver, BM and thymus revealing a requirement for Gata2 in embryonic hematopoiesis (Tsai et al., 1994). Besides the embryonic lethality of Gata2-null embryos, the number and function of hematopoietic stem and progenitor cells from germline heterozygous Gata2 mutant mice at E10.5–E12 is impaired (Ling et al., 2004).
Both in human and mouse embryos, Gata2 is expressed in a specialized endothelial cell population called hemogenic endothelium (HE) and in the first transplantable hematopoietic stem cells (HSCs) that differentiate from HE (Marshall et al., 1999; Yokomizo and Dzierzak, 2010; Eich et al., 2018; Vink et al., 2020). Conditional deletion of Gata2 in HE cells resulted in reduced hematopoietic cluster formation in the embryo and long-term repopulating HSCs were not formed. Conditional deletion of Gata2 in HSCs induced apoptosis indicating that GATA2 is required both for HSC generation and maintenance (de Pater et al., 2013).
Gata2 expression is regulated by the enhancer activity of multiple conserved cis-regulatory elements. The disruption of the +9.5 element of Gata2 impaired vascular integrity and formation of HSCs from HE in the mouse embryo (Lim et al., 2012; Gao et al., 2013).
Although both number and functionality of HSCs were reduced in embryonic Gata2 haploinsufficiency, it is yet to be discovered whether and how the propensity for MDS/AML observed in GATA2 haploinsufficiency patients is influenced by these early embryonic functions.
The Function of GATA2 in Adult Bone Marrow Hematopoiesis
The function of GATA2 in adult hematopoiesis is still abstruse. In BM, Gata2 is highly expressed in HSCs and downregulated during lineage commitment (Akashi et al., 2000; Miyamoto et al., 2002; Guo et al., 2013). HSCs in the BM of Gata2+/− mice are impaired in number and functionality as shown by serial transplantation assays (Rodrigues et al., 2005; Guo et al., 2013). In addition, Gata2-heterozygosity in BM HSCs is associated with a decreased proliferation ability together with increased quiescence and apoptosis (Ling et al., 2004; Rodrigues et al., 2005). Moreover, Gata2 haploinsufficiency reduces the function of granulocyte-macrophage progenitors but not of other myeloid committed progenitors (Rodrigues et al., 2008). However, Gata2+/− mice do not develop MDS/AML. This makes it difficult to study the contribution of GATA2 haploinsufficiency to leukemic progression in these models.
On the other hand, Gata2 overexpression results in the self-renewal of myeloid progenitors and blocks lymphoid differentiation in mouse BM (Nandakumar et al., 2015). In addition, overexpression of GATA2 in human ES cells (hESC) promotes proliferation in hESCs, but quiescence in hESC-derived HSCs (Zhou et al., 2019). Furthermore, increased GATA2 expression is also observed in adult and pediatric AML patients with poor prognosis (Ayala et al., 2009; Luesink et al., 2012; Vicente et al., 2012; Menendez-Gonzalez et al., 2019). These findings indicate that, next to its tumor suppressor role, GATA2 might act as an oncogene when overexpressed.
Genome Editing: A Cure for GATA2 Haploinsufficiencies?
GATA2 Repair Strategies
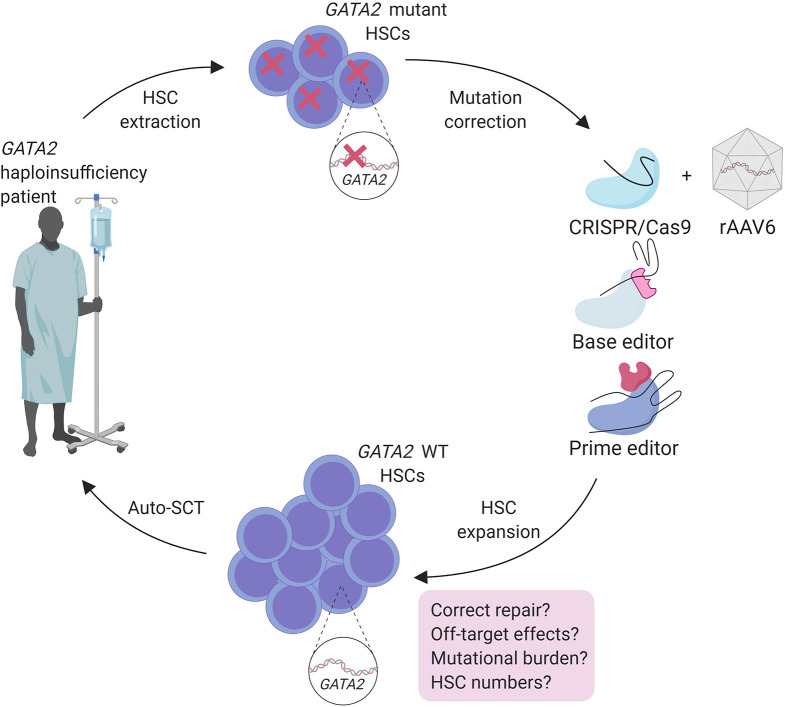
Allo-SCT is a powerful approach to treat malignancies in GATA2 haploinsufficiency patients (Simonis et al., 2018; van Lier et al., 2020). However, finding a matched donor and TRM compromises the use of allo-SCT and is therefore not suitable before the onset of malignancy (Bogaert et al., 2020). Regulation of GATA2 expression is crucial in HSCs and in leukemia predisposition. This makes overexpression of WT GATA2 using lenti-viral transgenic approaches not suitable as gene therapy method. An auto-SCT approach, after ex vivo correction of the underlying patient specific GATA2 mutation by genome editing tools is possibly a more effective treatment option for these patients (Figure 2).
Figure 2.
Treatment strategy for genome engineering of autologous HSC of GATA2 haploinsufficiency patients with safety considerations.
Genome editing, since it was pioneered in the previous century, is developing meteorically as a revolutionary therapeutic tool for genetic defects, including hematological disorders (Xie et al., 2014; Hoban et al., 2016; De Ravin et al., 2017; Orkin and Bauer, 2019) CRISPR/Cas9, a part of the bacterial acquired immune system, was adapted as a breakthrough genome engineering technology and has since been extensively used to engineer eukaryotic cells in basic research and holds great potential for gene therapy (Gasiunas et al., 2012; Jinek et al., 2012; Cong et al., 2013; Barrangou and Doudna, 2016). CRISPR/Cas9 mediated genome editing relies on sequence specific guide RNAs that assemble with Cas9 protein to create double strand breaks (DSBs) in the targeted sequence. DSBs activates cell intrinsic repair mechanisms if the cell is to undergo proliferation and repaired by one of two mechanisms: non-homologous end joining (NHEJ) in which random insertions/deletions (InDels) are introduced or homology-directed repair (HDR) which uses the other DNA strand as template to restore its original sequence. This system can be hijacked by providing an exogenous repair template containing any desired sequence. Because HDR is rare, a selection cassette can be inserted for positive selection of the desired repair (Doudna and Charpentier, 2014).
Because heterogeneity of mutations in GATA2 haploinsufficieny patients (https://www.ncbi.nlm.nih.gov/clinvar), these mutations would need to be restored at the endogenous locus, requiring HDR as repair mechanism. Therefore, optimizing an editing strategy by using large HDR donor templates that cover various GATA2 mutation regions found in patients or the whole gene, containing homologous regions covering several exons, could provide treatment for a substantial group of GATA2 patients. An efficient method for gene correction in HSCs with CRISPR/Cas9 and large HDR donor delivered by rAAV6 (adeno-associated viral vectors of serotype 6) was used to correct a HBB gene mutation causing sickle cell disease and has potential to correct GATA2 mutations in HSCs using the same strategy (Dever et al., 2016; DeWitt et al., 2016; Bak et al., 2018) (Figure 2).
Hurdles
GATA2 haploinsufficiencies result in a diminished number of HSCs in both embryonic and adult stages. Additionally, HDR mediated repair works with low efficiencies and studies showed that it is more efficient in hematopoietic progenitor cells rather than long-term repopulating HSCs (Genovese et al., 2014; Hoban et al., 2015). Together this implicates the biggest hurdle to treat GATA2 haploinsufficiency patients would be to obtain sufficient number of corrected HSCs for auto-SCT. An enrichment method, possibly a reporter-based selection followed by an ex vivo expansion of GATA2-corrected HSCs, could potentially solve this problem. For this purpose, small molecule drugs promoting ex vivo expansion of HSCs, like SR1 or UM171, could be used to obtain higher number of corrected HSCs prior to auto-SCT (Boitano et al., 2010; Fares et al., 2014).
Furthermore, in GATA2 haploinsufficiency patients, additional mutations in other genes could be the driver of leukemia which brings challenges to treat these patients by only correcting the mutant GATA2 allele. Therefore, a preliminary genetic screening for additional mutations should be compulsory in GATA2 haploinsufficiency patients to elucidate if correcting only the mutant GATA2 allele would eliminate the disease phenotype of the patient.
Another hurdle when using genome editing tools for clinical applications is the off-target effects (OTEs) that might occur in undesired parts of the DNA. Detection of OTEs with whole genome sequencing are often challenging due to high background of random reads in combination with low sequence depth (<10-fold) (Kim et al., 2015). More screening strategies for OTEs, like GUIDE-seq (Genome wide, Unbiased Identification of DSB Enabled by sequencing), CIRCLE-seq (Circularization for in vitro Reporting of Cleavage Effects) and DISCOVER-seq (Discovery of in situ Cas Off-targets and VERification by Sequencing), are shown to overcome these obstacles and could be used to efficiently identify OTEs that might result from GATA2-editing strategy before its clinical translation (Tsai et al., 2015, 2017; Wienert et al., 2019).
Prospects
Fortunately, recent improvements of CRISPR/Cas9 genome editing may overcome some of these hurdles for patient applications. Base editing methods are developed by the addition of enzymes to Cas9 to provide single base pair changes without making DSBs (Komor et al., 2016; Gaudelli et al., 2017). Although base editing can correct point mutations that are also found in GATA2 patients, the off-target effects caused by the broad activity of cytidine deaminases used in this method should be considered carefully (Zuo et al., 2019; Yu et al., 2020). More recently Anzalone et al. (2019) described prime editing that introduces specific insertions, deletions and point mutations to a variety of genomic regions with high efficiency without DSBs. Prime editing was successfully used in human cells to correct mutations that cause sickle cell disease and Tay-Sachs disease and only 1–10% of prime-edited cells are found to have unwanted off-target InDels throughout the genome (Anzalone et al., 2019). These recent advances in genome editing techniques anticipate the improvement of a safer and more efficient correction of the patient mutations in HSCs prior to auto-SCT, and should be considered for the treatment of GATA2 haploinsufficiencies (Figure 2).
Currently, the minimum level of donor chimerism necessary to reverse the disease phenotype in GATA2 haploinsufficiency patients remains unclear (Hickstein, 2018). For sickle cell disease however, it was shown that clinical benefits might be observed when as few as 2–5 HSCs are engrafted (Walters et al., 2001; DeWitt et al., 2016). Interestingly, an asymptomatic germline GATA2 mutant individual acquired a somatic mutation reversing the harmful GATA2 mutation. This resulted in a selective advantage of the corrected HSCs and prevented from developing malignancy (Catto et al., 2020). Together this implicates having a few mutation-corrected HSCs might already have clinical significance for GATA2 haploinsufficiency patients.
CRISPR/Cas9 technology has been approved in patient treatment for various types of malignancies including hematological diseases (https://clinicaltrials.gov). Currently, clinical trials are performed where CRISPR/Cas9 is used to remove erythroid expression of the fetal hemoglobin repressor BCL11A in the treatment of hemaglobinopathies, implicating a highly promising potential for genome editing to treat various hematological disorders (Orkin and Bauer, 2019; The Lancet Haematology, 2019).
Careful consideration of possible challenges discussed for GATA2 haploinsufficiency patients could lead to a beneficial clinical translation of genome editing to treat these patients in the near future.
Discussion
Although GATA2 haploinsufficiency depletes the HSC compartment in humans and mice, the function of GATA2 haploinsufficiency in MDS/AML progression is poorly understood. A possibility could be that GATA2 haploinsufficiency provides a fertile ground for the emergence of additional mutations in HSCs and these acquired mutations promote leukemogenesis. Evidence that support this hypothesis is the inconsistent penetrance of leukemia in GATA2 haploinsufficiency patients that cannot be explained solely by the mutations in the GATA2 locus and MDS/AML patients with germline GATA2 mutation presented with additional mutations which are linked to hematological malignancies (Wlodarski et al., 2016; Fisher et al., 2017; Yoshida et al., 2020). In order to understand the concept of fertile ground as a driver of MDS/AML in GATA2 deficiency syndromes, more fundamental research is needed to reveal the clonal origin (embryonic and/or adult) of leukemogenic driver mutations to help us choose an appropriate time frame and strategy to treat these patients using genome editing. If leukemic driver mutations arise early during hematopoietic development, targeting leukemic clones will be challenging.
in vivo Gata2+/− models have not developed an MDS/AML phenotype (Ling et al., 2004; Rodrigues et al., 2005). This could be due to differences governing HSC mechanisms in these models or due to differences in lifespan, infection status, genetic background or a combination of these factors. Perhaps aged Gata2+/− models could provide more insight, since this would challenge the HSC compartment and increase the chances of additional events that would promote leukemogenesis to occur.
Base editing and prime editing are the recent promising and rigorous refinements of genome editing technologies which could provide and improve a patient specific mutation correction for GATA2 mutations or any other gene mutations that predispose to hematological malignancies when potentials and risks of these tools are tested sufficiently prior to the actual patient treatments. In addition to their potential for gene therapy discussed in this review, CRISPR base and prime editing technologies are also fantastic tools for basic research to introduce additional predicted leukemia driver mutations to HSCs in GATA2 haploinsufficiency models in order to identify their potential role in malignant transformation.
Author Contributions
CK and EP wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. IP Touw and Dr. T Cupedo for careful reading of the manuscript. Figures are created with BioRender.com.
Footnotes
Funding. EP was supported by grant SK_10321 (Dutch Cancer foundation/Alpe d'Huzes).
References
- Akashi K., Reya T., Dalma-Weiszhausz D., Weissman I. L. (2000). Lymphoid precursors. Curr. Opin. Immunol. 12, 144–150. 10.1016/S0952-7915(99)00064-3 [DOI] [PubMed] [Google Scholar]
- Alfayez M., Wang S. A., Bannon S. A., Kontoyiannis D. P., Kornblau S. M., Orange J. S., et al. (2019). Myeloid malignancies with somatic GATA2 mutations can be associated with an immunodeficiency phenotype. Leuk. Lymphoma 60, 2025–2033. 10.1080/10428194.2018.1551535 [DOI] [PubMed] [Google Scholar]
- Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala R. M., Martínez-López J., Albízua E., Diez A., Gilsanz F. (2009). Clinical significance of Gata-1, Gata-2, EKLF, and c-MPL expression in acute myeloid leukemia. Am. J. Hematol. 84, 79–86. 10.1002/ajh.21332 [DOI] [PubMed] [Google Scholar]
- Bak R. O., Dever D. P., Porteus M. H. (2018). CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nat. Protoc. 13, 358–376. 10.1038/nprot.2017.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Doudna J. A. (2016). Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941. 10.1038/nbt.3659 [DOI] [PubMed] [Google Scholar]
- Bogaert D. J., Laureys G., Naesens L., Mazure D., De Bruyne M., Hsu A. P., et al. (2020). GATA2 deficiency and haematopoietic stem cell transplantation: challenges for the clinical practitioner. Br. J. Haematol. 188, 768–773. 10.1111/bjh.16247 [DOI] [PubMed] [Google Scholar]
- Boitano A. E., Wang J., Romeo R., Bouchez L. C., Parker A. E., Sutton S. E., et al. (2010). Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329, 1345–1348. 10.1126/science.1191536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catto L. F. B., Borges G., Pinto A. L., Cl,é D. V., Chahud F., et al. (2020). Somatic genetic rescue in hematopoietic cells in GATA2 deficiency. Blood 136, 1002–1005. 10.1182/blood.2020005538 [DOI] [PubMed] [Google Scholar]
- Collin M., Dickinson R., Bigley V. (2015). Haematopoietic and immune defects associated with GATA2 mutation. Br. J. Haematol. 169, 173–187. 10.1111/bjh.13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. R. (2018). 5 - bone marrow failure syndromes, in Hematopathology, 3rd Edn. ed. E. D. Hsi (Philadelphia, PA: Elsevier; ), 167–183.e161. 10.1016/B978-0-323-47913-4.00005-7 [DOI] [Google Scholar]
- de Pater E., Kaimakis P., Vink C. S., Yokomizo T., Yamada-Inagawa T. R., et al. (2013). Gata2 is required for HSC generation and survival. J. Exp. Med. 210, 2843–2850. 10.1084/jem.20130751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ravin S. S., Li L., Wu X., Choi U., Allen C., Koontz S., et al. (2017). CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci. Transl. Med. 9:aah3480. 10.1126/scitranslmed.aah3480 [DOI] [PubMed] [Google Scholar]
- Dever D. P., Bak R. O., Reinisch A., Camarena J., Washington G., Nicolas C. E., et al. (2016). CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389. 10.1038/nature20134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt M. A., Magis W., Bray N. L., Wang T., Berman J. R., Urbinati F., et al. (2016). Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 8:360ra134. 10.1126/scitranslmed.aaf9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R. E., Griffin H., Bigley V., Reynard L. N., Hussain R., Haniffa M., et al. (2011). Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118, 2656–2658. 10.1182/blood-2011-06-360313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal I., Vulliamy T. (2010). Inherited bone marrow failure syndromes. Haematologica 95, 1236–1240. 10.3324/haematol.2010.025619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadieu J., Lamant M., Fieschi C., de Fontbrune F. S., Caye A., Ouachee M., et al. (2018). Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica 103, 1278–1287. 10.3324/haematol.2017.181909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Eich C., Arlt J., Vink C. S., Solaimani Kartalaei P., Kaimakis P., Mariani S. A., et al. (2018). In vivo single cell analysis reveals Gata2 dynamics in cells transitioning to hematopoietic fate. J. Exp. Med. 215, 233–248. 10.1084/jem.20170807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Felsenfeld G. (1989). The erythroid-specific transcription factor Eryf1: a new finger protein. Cell 58, 877–885. 10.1016/0092-8674(89)90940-9 [DOI] [PubMed] [Google Scholar]
- Fares I., Chagraoui J., Gareau Y., Gingras S., Ruel R., Mayotte N., et al. (2014). Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 345, 1509–1512. 10.1126/science.1256337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasan A., Eder C., Haferlach C., Grossmann V., Kohlmann A., Dicker F., et al. (2013). GATA2 mutations are frequent in intermediate-risk karyotype AML with biallelic CEBPA mutations and are associated with favorable prognosis. Leukemia 27, 482–485. 10.1038/leu.2012.174 [DOI] [PubMed] [Google Scholar]
- Fisher K. E., Hsu A. P., Williams C. L., Sayeed H., Merritt B. Y., Elghetany M. T., et al. (2017). Somatic mutations in children with GATA2-associated myelodysplastic syndrome who lack other features of GATA2 deficiency. Blood Adv. 1, 443–448. 10.1182/bloodadvances.2016002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi K. A., Townsley D. M., Hsu A. P., Arthur D. C., Zerbe C. S., Cuellar-Rodriguez J., et al. (2015). GATA2 deficiency-associated bone marrow disorder differs from idiopathic aplastic anemia. Blood 125, 56–70. 10.1182/blood-2014-06-580340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Johnson K. D., Chang Y. I., Boyer M. E., Dewey C. N., Zhang J., et al. (2013). Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J. Exp. Med. 210, 2833–2842. 10.1084/jem.20130733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, E2579–2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli N. M., Komor A. C., Rees H. A., Packer M. S., Badran A. H., Bryson D. I., et al. (2017). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese P., Schiroli G., Escobar G., Tomaso T. D., Firrito C., Calabria A., et al. (2014). Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240. 10.1038/nature13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Luc S., Marco E., Lin T. W., Peng C., Kerenyi M. A., et al. (2013). Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 13, 492–505. 10.1016/j.stem.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. N., Chong C. E., Carmichael C. L., Wilkins E. J., Brautigan P. J., Li X. C., et al. (2011). Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 43, 1012–1017. 10.1038/ng.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickstein D. (2018). HSCT for GATA2 deficiency across the pond. Blood 131, 1272–1274. 10.1182/blood-2018-02-826461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban M. D., Cost G. J., Mendel M. C., Romero Z., Kaufman M. L., Joglekar A. V., et al. (2015). Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125, 2597–2604. 10.1182/blood-2014-12-615948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban M. D., Lumaquin D., Kuo C. Y., Romero Z., Long J., Ho M., et al. (2016). CRISPR/Cas9-mediated correction of the sickle mutation in human CD34+ cells. Mol. Ther. 24, 1561–1569. 10.1038/mt.2016.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H. A., Lin Y. C., Kuo Y. Y., Chou W. C., Lin C. C., Liu C. Y., et al. (2015). GATA2 mutations in patients with acute myeloid leukemia-paired samples analyses show that the mutation is unstable during disease evolution. Ann. Hematol. 94, 211–221. 10.1007/s00277-014-2208-8 [DOI] [PubMed] [Google Scholar]
- Hsu A. P., Johnson K. D., Falcone E. L., Sanalkumar R., Sanchez L., Hickstein D. D., et al. (2013). GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood 121, 3830–3837. 10.1182/blood-2012-08-452763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A. P., Sampaio E. P., Khan J., Calvo K. R., Lemieux J. E., Patel S. Y., et al. (2011). Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118, 2653–2655. 10.1182/blood-2011-05-356352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Bae S., Park J., Kim E., Kim S., Yu H. R., et al. (2015). Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243. 10.1038/nmeth.3284 [DOI] [PubMed] [Google Scholar]
- Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. C., Hosoya T., Brandt W., Ku C. J., Hosoya-Ohmura S., Camper S. A., et al. (2012). Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J. Clin. Invest. 122, 3705–3717. 10.1172/JCI61619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K. W., Ottersbach K., van Hamburg J. P., Oziemlak A., Tsai F. Y., Orkin S. H., et al. (2004). GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200, 871–882. 10.1084/jem.20031556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesink M., Hollink I. H. V. H, van der Velden Knops R. H., Boezeman J. B., de Haas V., et al. (2012). High GATA2 expression is a poor prognostic marker in pediatric acute myeloid leukemia. Blood 120, 2064–2075. 10.1182/blood-2011-12-397083 [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C. J., Moore R. L., Thorogood P., Brickell P. M., Kinnon C., Thrasher A., et al. (1999). Detailed characterization of the human aorta-gonad-mesonephros region reveals morphological polarity resembling a hematopoietic stromal layer. Dev. Dyn. 215, 139–147. [DOI] [PubMed] [Google Scholar]
- McReynolds L. J., Calvo K. R., Holland S. M. (2018). Germline GATA2 mutation and bone marrow failure. Hematol. Oncol. Clin. North Am. 32, 713–728. 10.1016/j.hoc.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Gonzalez J. B., Vukovic M., Abdelfattah A., Saleh L., Almotiri A., Thomas L. A., et al. (2019). Gata2 as a crucial regulator of stem cells in adult hematopoiesis and acute myeloid leukemia. Stem Cell Rep. 13, 291–306. 10.1016/j.stemcr.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi N., Ohta J., Suwabe N., Nakauchi H., Ishihara H., Hayashi N., et al. (1998). Alternative promoters regulate transcription of the mouse GATA-2 gene. J. Biol. Chem. 273, 3625–3634. 10.1074/jbc.273.6.3625 [DOI] [PubMed] [Google Scholar]
- Miyamoto T., Iwasaki H., Reizis B., Ye M., Graf T., Weissman I. L., et al. (2002). Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3, 137–147. 10.1016/S1534-5807(02)00201-0 [DOI] [PubMed] [Google Scholar]
- Nandakumar S. K., Johnson K., Throm S. L., Pestina T. I., Neale G., Persons D., et al. (2015). Low-level GATA2 overexpression promotes myeloid progenitor self-renewal and blocks lymphoid differentiation in mice. Exp. Hematol. 43, 565–577.e561–e510. 10.1016/j.exphem.2015.04.002 [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Bauer D. E. (2019). Emerging genetic therapy for sickle cell disease. Annu. Rev. Med. 70, 257–271. 10.1146/annurev-med-041817-125507 [DOI] [PubMed] [Google Scholar]
- Ostergaard P., Simpson M. A., Connell F. C., Steward C. G., Brice G., Woollard W. J., et al. (2011). Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat. Genet. 43, 929–931. 10.1038/ng.923 [DOI] [PubMed] [Google Scholar]
- Pan X., Minegishi N., Harigae H., Yamagiwa H., Minegishi M., Akine Y., et al. (2000). Identification of human GATA-2 gene distal IS exon and its expression in hematopoietic stem cell fractions. J. Biochem. 127, 105–112. 10.1093/oxfordjournals.jbchem.a022570 [DOI] [PubMed] [Google Scholar]
- Pasquet M., Bellanné-Chantelot C., Tavitian S., Prade N., Beaupain B., Larochelle O., et al. (2013). High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood 121, 822–829. 10.1182/blood-2012-08-447367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping N., Sun A., Song Y., Wang Q., Yin J., Cheng W., et al. (2017). Exome sequencing identifies highly recurrent somatic GATA2 and CEBPA mutations in acute erythroid leukemia. Leukemia 31, 195–202. 10.1038/leu.2016.162 [DOI] [PubMed] [Google Scholar]
- Rodrigues N. P., Boyd A. S., Fugazza C., May G. E., Guo Y., Tipping A. J., et al. (2008). GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood 112, 4862–4873. 10.1182/blood-2008-01-136564 [DOI] [PubMed] [Google Scholar]
- Rodrigues N. P., Janzen V., Forkert R., Dombkowski D. M., Boyd A. S., Orkin S. H., et al. (2005). Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 106, 477–484. 10.1182/blood-2004-08-2989 [DOI] [PubMed] [Google Scholar]
- Sekhar M., Pocock R., Lowe D., Mitchell C., Marafioti T., Dickinson R., et al. (2018). Can somatic GATA2 mutation mimic germ line GATA2 mutation? Blood Adv. 2, 904–908. 10.1182/bloodadvances.2017012617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis A., Fux M., Nair G., Mueller N. J., Haralambieva E., Pabst T., et al. (2018). Allogeneic hematopoietic cell transplantation in patients with GATA2 deficiency-a case report and comprehensive review of the literature. Ann. Hematol. 97, 1961–1973. 10.1007/s00277-018-3388-4 [DOI] [PubMed] [Google Scholar]
- Spinner M. A., Sanchez L. A., Hsu A. P., Shaw P. A., Zerbe C. S., Calvo K. R., et al. (2014). GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood 123, 809–821. 10.1182/blood-2013-07-515528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Lancet Haematology (2019). CRISPR-Cas9 gene editing for patients with haemoglobinopathies. Lancet Haematol. 6:e438. 10.1016/S2352-3026(19)30169-3 [DOI] [PubMed] [Google Scholar]
- Theis F., Corbacioglu A., Gaidzik V. I., Paschka P., Weber D., Bullinger L., et al. (2016). Clinical impact of GATA2 mutations in acute myeloid leukemia patients harboring CEBPA mutations: a study of the AML study group. Leukemia 30, 2248–2250. 10.1038/leu.2016.185 [DOI] [PubMed] [Google Scholar]
- Tien F. M., Hou H. A., Tsai C. H., Tang J. L., Chiu Y. C., Chen C. Y., et al. (2018). GATA2 zinc finger 1 mutations are associated with distinct clinico-biological features and outcomes different from GATA2 zinc finger 2 mutations in adult acute myeloid leukemia. Blood Cancer J. 8:87. 10.1038/s41408-018-0123-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley D. M., Hsu A., Dumitriu B., Holland S. M., Young N. S. (2012). Regulatory mutations in GATA2 associated with aplastic anemia. Blood 120, 3488–3488. 10.1182/blood.V120.21.3488.348822955925 [DOI] [Google Scholar]
- Tsai F. Y., Keller G., Kuo F. C., Weiss M., Chen J., Rosenblatt M., et al. (1994). An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226. 10.1038/371221a0 [DOI] [PubMed] [Google Scholar]
- Tsai S. Q., Nguyen N. T., Malagon-Lopez J., Topkar V. V., Aryee M. J., Joung J., et al. (2017). CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614. 10.1038/nmeth.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Q., Zheng Z., Nguyen N. T., Liebers M., Topkar V. V., Thapar V., et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197. 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lier Y. F., de Bree G. J., Jonkers R. E., Roelofs J., Ten Berge I. J. M., et al. (2020). Allogeneic hematopoietic cell transplantation in the management of GATA2 deficiency and pulmonary alveolar proteinosis. Clin. Immunol. 218:108522. 10.1016/j.clim.2020.108522 [DOI] [PubMed] [Google Scholar]
- Vicente C., Conchillo A., García-Sánchez M. A., Odero M. D. (2012). The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit. Rev. Oncol. Hematol. 82, 1–17. 10.1016/j.critrevonc.2011.04.007 [DOI] [PubMed] [Google Scholar]
- Vink C. S., Calero-Nieto F. J., Wang X., Maglitto A., Mariani S. A., Jawaid W., et al. (2020). Iterative single-cell analyses define the transcriptome of the first functional hematopoietic stem cells. Cell Rep. 31:107627. 10.1016/j.celrep.2020.107627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M. C., Patience M., Leisenring W., Rogers Z. R., Aquino V. M., Buchanan G. R., et al. (2001). Stable mixed hematopoietic chimerism after bone marrow transplantation for sickle cell anemia. Biol. Blood Marrow Transplant 7, 665–673. 10.1053/bbmt.2001.v7.pm11787529 [DOI] [PubMed] [Google Scholar]
- Wienert B., Wyman S. K., Richardson C. D., Yeh C. D., Akcakaya P., Porritt M. J., et al. (2019). Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286–289. 10.1101/469635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. B., Link D. C., Mason P. J., Bessler M. (2014). Inherited bone marrow failure syndromes in adolescents and young adults. Ann. Med. 46, 353–363. 10.3109/07853890.2014.915579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarski M. W., Collin M., Horwitz M. S. (2017). GATA2 deficiency and related myeloid neoplasms. Semin. Hematol. 54, 81–86. 10.1053/j.seminhematol.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarski M. W., Hirabayashi S., Pastor V., Star,ý J., Hasle H., Masetti R., et al. (2016). Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood 127, 1387–1397; quiz 1518. 10.1182/blood-2015-09-669937 [DOI] [PubMed] [Google Scholar]
- Xie F., Ye L., Chang J. C., Beyer A. I., Wang J., Muench M. O., et al. (2014). Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 24, 1526–1533. 10.1101/gr.173427.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T., Dzierzak E. (2010). Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development 137, 3651–3661. 10.1242/dev.051094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Tanase-Nakao K., Shima H., Shirai R., Yoshida K., Osumi T., et al. (2020). Prevalence of germline GATA2 and SAMD9/9L variants in paediatric haematological disorders with monosomy 7. Br. J. Haematol. [Epub ahead of print]. 10.1111/bjh.17006 [DOI] [PubMed] [Google Scholar]
- Yu Y., Leete T. C., Born D. A., Young L., Barrera L. A., Lee S. J., et al. (2020). Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun. 11:2052. 10.1038/s41467-020-15887-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang Y., Chen B., Dong Y., Zhang Y., Mao B., et al. (2019). Overexpression of GATA2 enhances development and maintenance of human embryonic stem cell-derived hematopoietic stem cell-like progenitors. Stem Cell Rep. 13, 31–47. 10.1016/j.stemcr.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., et al. (2019). Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292. 10.1126/science.aav9973 [DOI] [PMC free article] [PubMed] [Google Scholar]