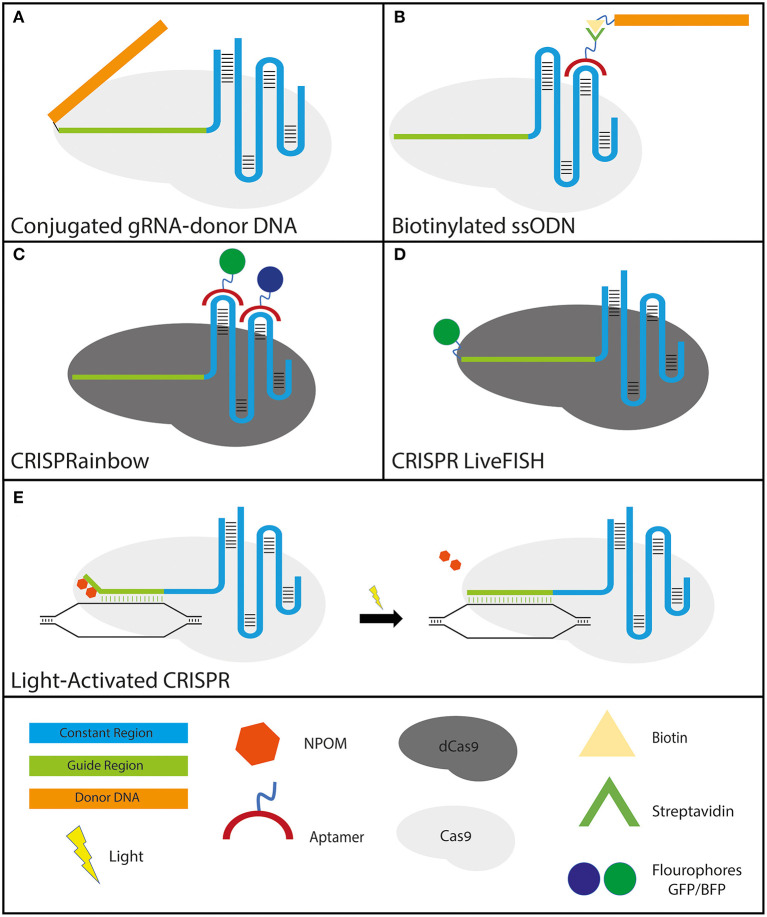
Figure 3.
Various applications of engineered gRNAs. (A,B)–gRNA modifications to improve HDR: (A) crRNA-donor DNA conjugate. The donor DNA is fused to the 5′ end of the guide region. (B) sgRNA molecule with streptavidin-binding aptamers that attach to either the tetraloop or stem-loop 2 (the two loops protruding from the Cas9 molecule). The formulation has the donor ssODN bound to a biotin molecule that binds the streptavidin tightly to ensure the proximity of the donor DNA to the break site. (C,D)–gRNA modifications that utilize CRISPR-dCas9 specificity for high-resolution cellular imaging: (C) sgRNA molecule with fluorophore-bound aptamers binding to either the tetraloop or stem-loop 2 (for the same reason as mentioned above). GFP and BFP were shown solely as examples since CRISPRainbow covers the full spectrum of combinations. (D) CRISPR LiveFISH method utilizes crRNAs fused to a fluorophore at the 5′ end to actuate live intracellular staining without the need for cellular fixation. (E) Light-activated CRISPR to allow for control over synchronous editing across a cell population. Photocaging with light-sensitive 6′-nitropiperonyloxymethyl (NPOM) thymidine modifications on the distal portion of the guide region prevents the gRNA from binding completely to its DNA target. Following exposure to light, the NPOM modifications are released and complete binding and subsequent editing commence.