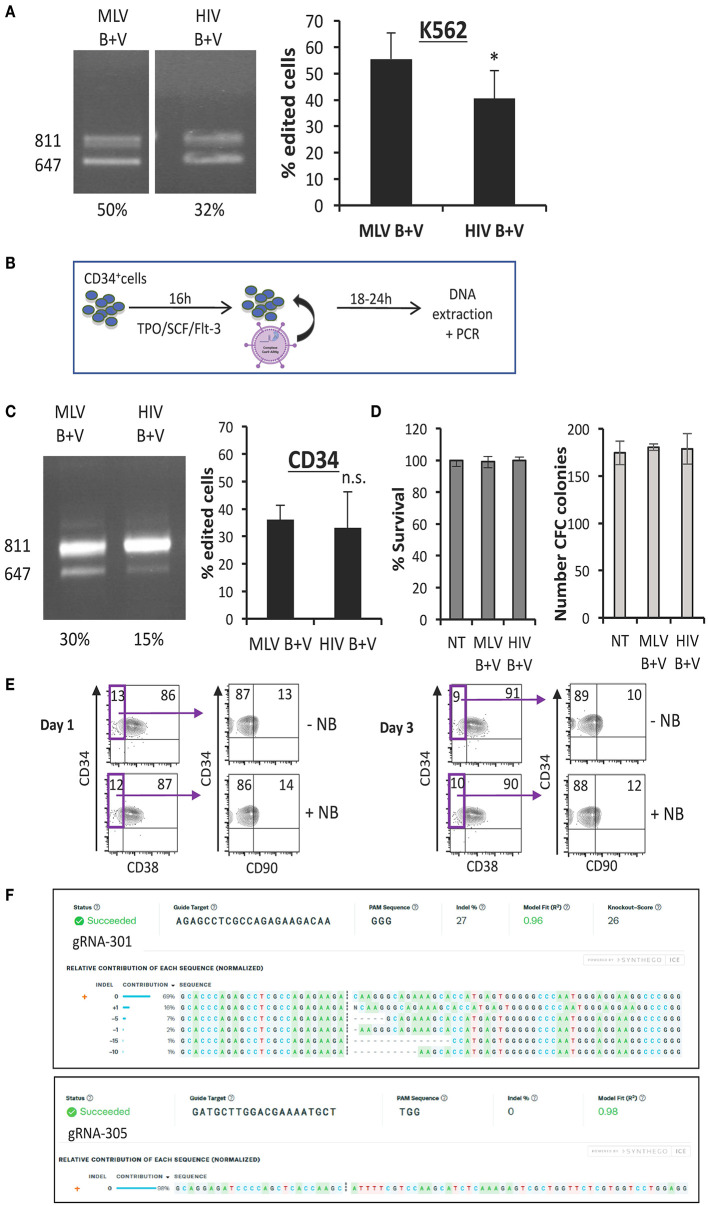
Figure 5.
Nanoblades permit efficient gene-editing in human K562 cells and CD34+ cells. (A) K562 cells transduced with MLV- or HIV-based nanoblades co-pseudotyped with BaEV- and VSVG-gps (B + V). Electrophoresis gel of the PCR product using WASFw/WASRv primers with the % gene-editing indicated for each lane (left) and summarizing graph showing percentage of gene editing (right) (means ± SD; (MLV B + V n = 7, HIV B + V n = 6; student t-test, *p < 0.05). (B) Schematic presentation of protocol used for CD34+ cells. CD34+ cells were activated with cytokines (TPO/SCF/Flt-3) for 16 h, then nanoblades were added and DNA extraction was performed 24 h post-transduction. (C) CD34+ cells transduced with MLV or HIV nanoblades co-pseudotyped with BaEV- and VSVG-gps (B + V). Electrophoresis gel of the PCR product using WASFw/WASRv primers with the % gene-editing indicated for each lane (left) and summarizing graph showing percentage of gene editing (right) (MLV B + V n = 7, HIV B + V n = 6; student t-test, n.s., not significant). (D) Survival of CD34+ cells 72 h post-incubation with nanoblades as determined by propidium iodide staining and flow cytometry analysis (left graph) and CFC frequencies of CD34+ cells incubated for 16 h with or without MLV or HIV nanoblades and upon differentiation for 14 days into myeloid lineages (means ± SD MLV B + V n = 3, HIV B + V n = 3; n.s., not significant). (E) Surface staining of CD34+ cells for CD38 and CD90 at day 1 or 3 post MLV-nanoblade incubation (+NB) compared to controls (–NB). These data are representative of n = 2. (F) Sequence decomposition of INDEL events. PCR indicated DSBs at gRNA 301 or 305 target loci simultaneously by exon 1 drop-out (647 bp band; Figure 2B). Sanger sequencing of these gRNA 301 or 305 loci in the residual 811 bp PCR was subjected to ICE analysis which revealed on-target INDEL frequencies at gRNA 301 or 305 target loci individually.