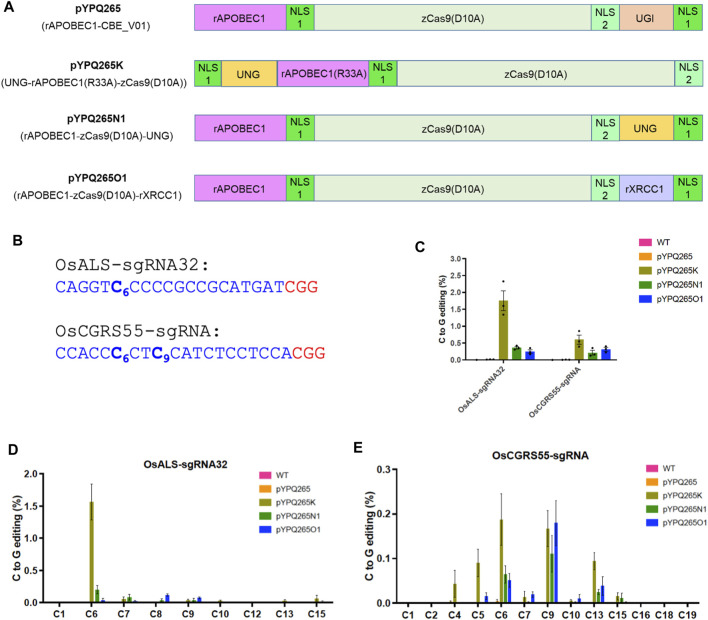
FIGURE 1.
Assessment of BE3 and three CGBEs in rice protoplasts. (A) Diagram of BE3 and three CGBEs. Note each nuclear localization single (NLS) is indicated by a green box. NLS 1 is a monopartite SV40 nuclear localization signal and NLS 2 is a bipartite nuclear localization signal of nucleoplasmin. Both NLS1 and NLS2 are recognized by importin α. (B) The target sites in the rice genome. The protospacer sequence is highlighted in blue and the PAM is highlighted in red. (C) NGS quantification of C-to-G editing by four base editors in rice protoplasts. For the wild type (WT) samples, sterile deionized water was used in protoplast transformation. (D) NGS analysis of C-to-G editing windows by different base editors at the OsALS-sgRNA32 site. (E) NGS analysis of the C-to-G editing windows by different base editors at the OsCGRS55-sgRNA site. The error bars represent standard errors of three biological replicates.