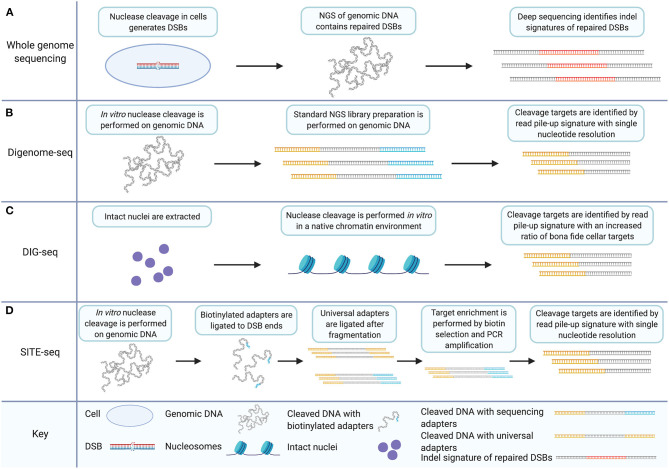
Figure 1.
In vitro off-target detection methods based on WGS utilize target enrichment strategies that facilitate data analysis and improve signal to noise ratios. (A) WGS for CRISPR/Cas9 off-target detection involves nuclease induction of DSBs in cells. Deep sequencing of genomic DNA captures all sequence including indels and genomic rearrangements introduced during DSB repair. (B) Digenome-seq involves extraction of genomic DNA and in vitro CRISPR/Cas9 treatment which precludes DSB repair. Samples are prepared by standard NGS methods where adapters (in yellow) are added. Cleavage targets are identified by a read pile-up signature with single-nucleotide resolution. (C) DIG-seq involves nuclei extraction. Nuclease cleavage is carried out in vitro in the native chromatin environment and libraries are prepared for NGS. Cleavage targets are identified by read pile-up, as in standard Digenome-seq. Intact chromatin reduces false discovery and increases the ratio of bona fide cellular targets identified. (D) SITE-Seq involves in vitro Cas9 digestion, as in Digenome-seq and DIG-seq. Biotinylated adapters (in blue) are then ligated to DSB ends followed by fragmentation and universal adapter ligation. Target enrichment is achieved by biotin selection and PCR amplification. As in Digenome-seq and DIG-seq, cleavage targets are identified by read pile-up during analysis. Created with Biorender.com.