Figure 3.
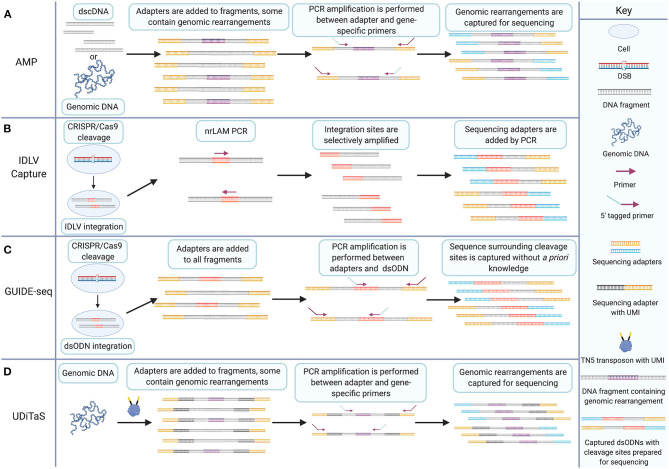
The GUIDE-seq target enrichment strategy combines IDLV capture and AMP for CRISPR/Cas9 cleavage detection without a priori knowledge. (A) AMP involves adding half-functional adapters (shown in yellow) to both ends of double-stranded cDNA or sheared genomic DNA. Fragments may contain genomic rearrangements that are amplified by PCR between a single anchored gene-specific primer site and a half-functional adapter. 5′ tags on primers enable addition of a second sequencing adapter to amplified target sites. Non-target sites do not get the additional adapter and are excluded from sequencing. (B) IDLV capture involves transfection of CRISPR/Cas9 and transduction of IDLV which is integrated into CRISPR/Cas9-induced DSBs during NHEJ, shown in red. nrLAM PCR selectively amplifies cleavage sites from the integrated sequence. Additional rounds of PCR add sequencing adapters (shown in blue and yellow) to the amplicons. (C) GUIDE-seq involves transfection of CRISPR/Cas9 and dsODN linkers (shown in red) that are incorporated into cleavage sites during NHEJ. Genomic DNA is fragmented, and half-functional universal adapters (shown in yellow) are added to all fragments. PCR amplification between dsODN and half-functional universal adapters using 5′ tagged primers enables selective amplification of sequence surrounding cleavage sites and the addition of a second adapter necessary for sequencing. (D) UDiTaS involves tagmentation of genomic DNA from nuclease-edited cells. Tagmentation fragments DNA and introduces unique molecular indices (UMIs) and adapters. Target enrichment is achieved by selective amplification of fragments between adapters and gene-specific sites. Genomic rearrangements can then be sequence using next-generation sequencing platforms. Created with Biorender.com.