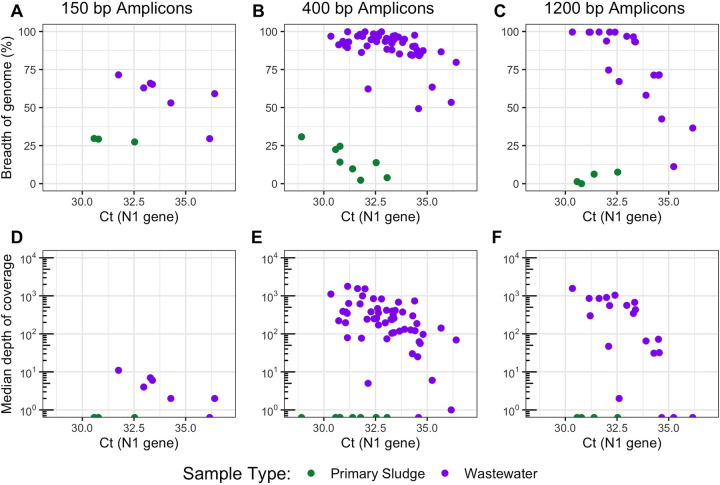
FIG 1.
SARS-CoV-2 whole-genome sequencing coverage results from three multiplex tiling PCR primer schemes, including breadth of genome coverage for 150-bp amplicons (A), 400-bp amplicons (B), and 1,200-bp amplicons (C), as well as the median depth of coverage across the genome for 150-bp amplicons (D), 400-bp amplicons (E), and 1,200-bp amplicons (F). The breadth of coverage represents the proportion of nucleotides in the SARS-CoV-2 reference genome (NCBI accession number MN908947.3) that are covered by at least one read, and the median depth of coverage represents the median number of reads mapped at each nucleotide position across the SARS-CoV-2 reference genome. Values are plotted versus the sample cycle threshold (CT) value for the U.S. CDC N1 assay, measured by reverse transcription-quantitative PCR (RT-qPCR) (see Text S1 in the supplemental material). Data points aligned with the x axis (plots D to F) had values of zero and could not be log transformed.