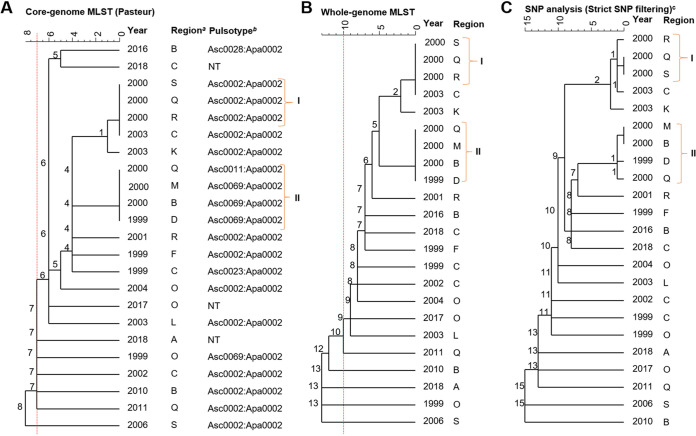
FIG 5.
Dendrograms (single linkage) representing the genetic relationship between L. monocytogenes isolates identified as L2-SL8-ST120-CT786 by using core genome multilocus sequence typing (cgMLST) (A), whole-genome MLST (wgMLST) (B), and single-nucleotide polymorphism (SNP) (C). Numbers on the branches indicate the allele/SNP differences between isolates. Isolates indicated with I are confirmed outbreak isolates. Those isolates indicated with II had low genetic diversity and occurred at approximately the same time as the outbreak isolates (I). a, regions within New Zealand have been anonymized; b, pulsotype assigned as a part of the PulseNet Aotearoa/New Zealand database; NT, not tested; c, SNP analysis was performed using the first clinical case (1999) for L2-SL8-ST120-CT786 as the reference. Red dotted lines represent cutoffs of 7 and 10 core and whole-genome allele differences, respectively (A and B), as previously proposed (6, 13).