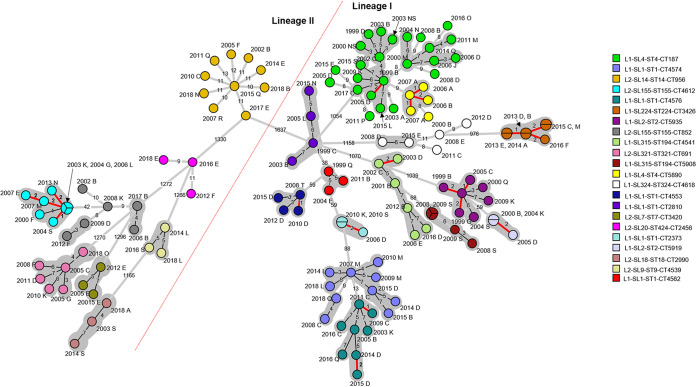
FIG 6.
Minimum spanning tree based on core genome multilocus sequence typing of all L. monocytogenes within core genome multilocus sequence types containing more than two isolates (excluding L2-SL8-ST120-CT786). Branch length scaling is logarithmic. The numbers of allele differences between isolates are shown on the branches. The year and region code are displayed next to each isolate. NS is used where a region was not stated. Isolates within a gray-shaded area share seven or less core genome allele differences between them, as previously proposed as a threshold (13). Isolates within a pie have zero core differences, and isolates that share 1 to 2 allele differences are highlighted with a red branch. A red dotted line separates lineages I and II.