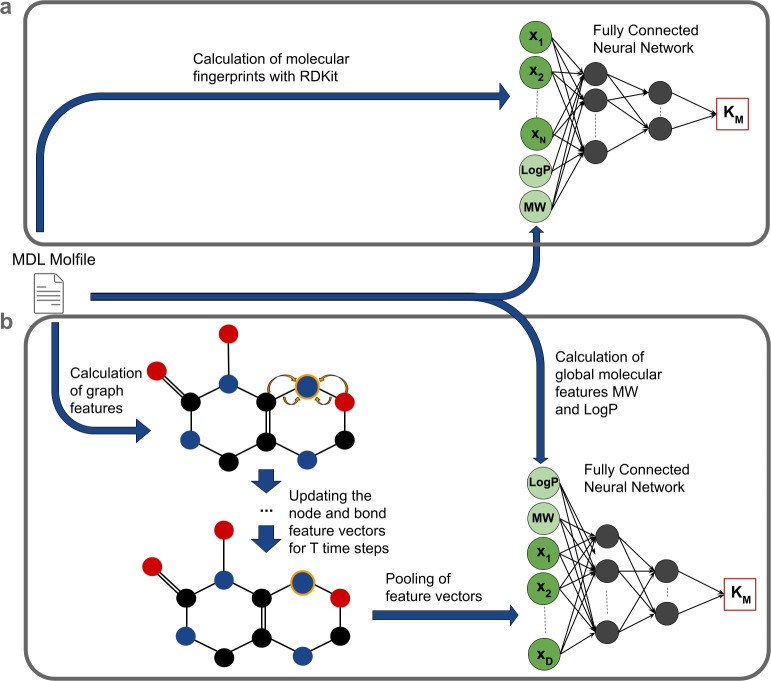
Fig 1. Model overview.
(a) Predefined molecular fingerprints. Molecular fingerprints are calculated from MDL Molfiles of the substrates and then passed through machine learning models like the FCNN together with 2 global features of the substrate, the MW and LogP. (b) GNN fingerprints. Node and edge feature vectors are calculated from MDL Molfiles and are then iteratively updated for T time steps. Afterwards, the feature vectors are pooled together into a single vector that is passed through an FCNN together with the MW and LogP. FCNN, fully connected neural network; GNN, graph neural network; LogP, octanol–water partition coefficient; MW, molecular weight.