Fig. 6 |. Primary probe synthesis.
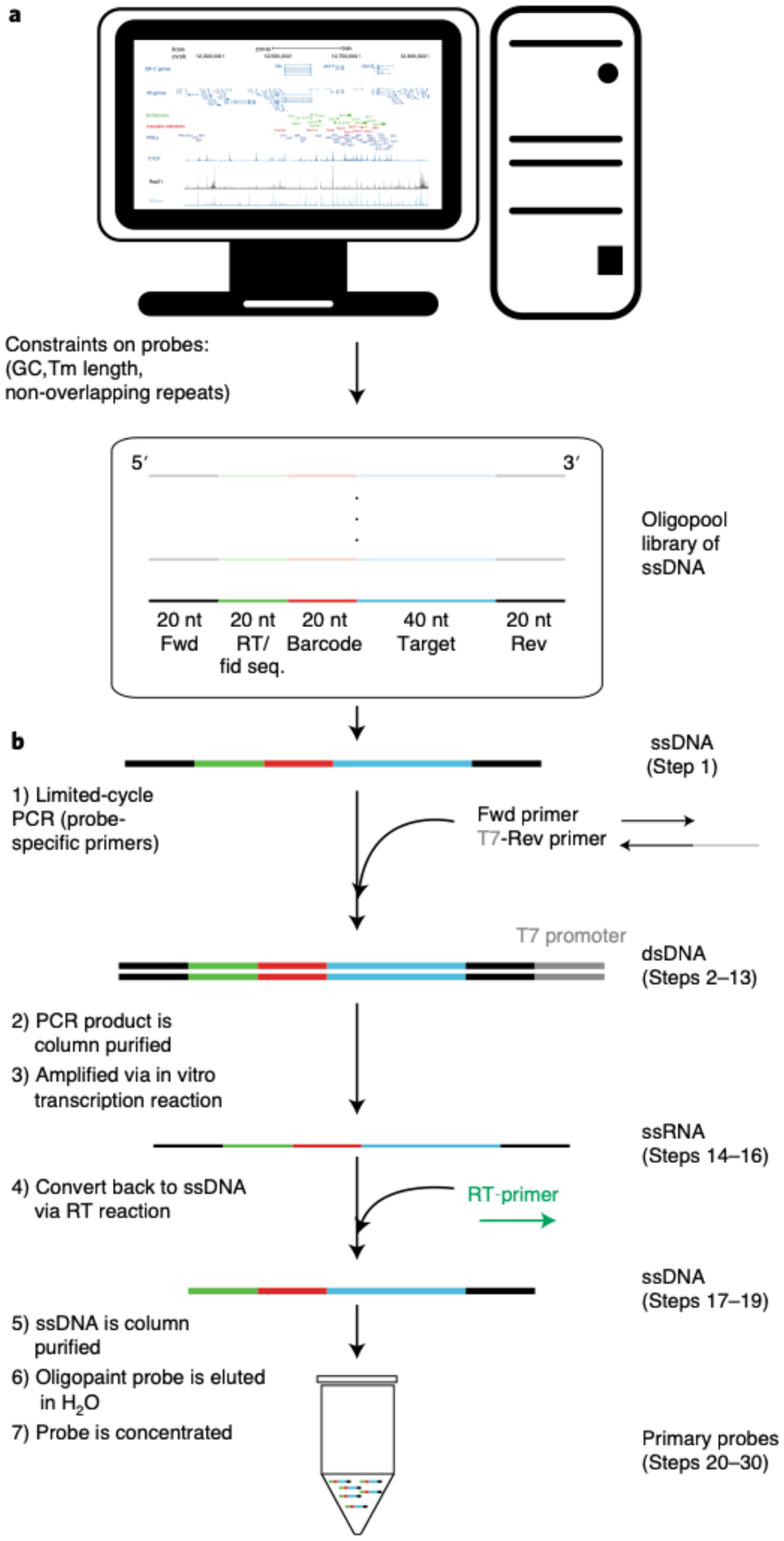
a, Computational design of the Oligopaint library. The region of interest is partitioned into genomic segments (e.g. 2 kb or 10 kb) and each segment is assigned a minimum of twenty 40-nt target sequences. The following are taken into consideration for the oligonucleotides: GC content melting temperature, homology to barcode sequences, and repetitive sequences. Our MATLAB scripts take into account these constraints and tile the oligonucleotides across a genomic region of interest in a uniform manner. Each primary probeset from the oligopool has a unique forward (Fwd) and reverse (Rev) index primer sequence and a 20-nt reverse transcription (RT) sequence (catcaacgccacgatcagct) that will be used for probe synthesis. Additionally, the RT sequence serves as the fiducial probe binding sequence (Fid seq.), which will be used for drift-correction after data collection. b, Schematic of the primary probe synthesis. Unique primers allow for the amplification specific primary probes via limited-cycle PCR. Following the PCR, the double-stranded DNA (dsDNA) undergoes an in vitro transcription reaction to generate a bulk number of single-stranded RNAs (ssRNAs), which are converted back to single stranded DNA via a reverse transcription reaction. The DNA is then column purified, and the primary probes are concentrated.
