Fig. 9 |. Microscope and fluidics setup for an automated experiment.
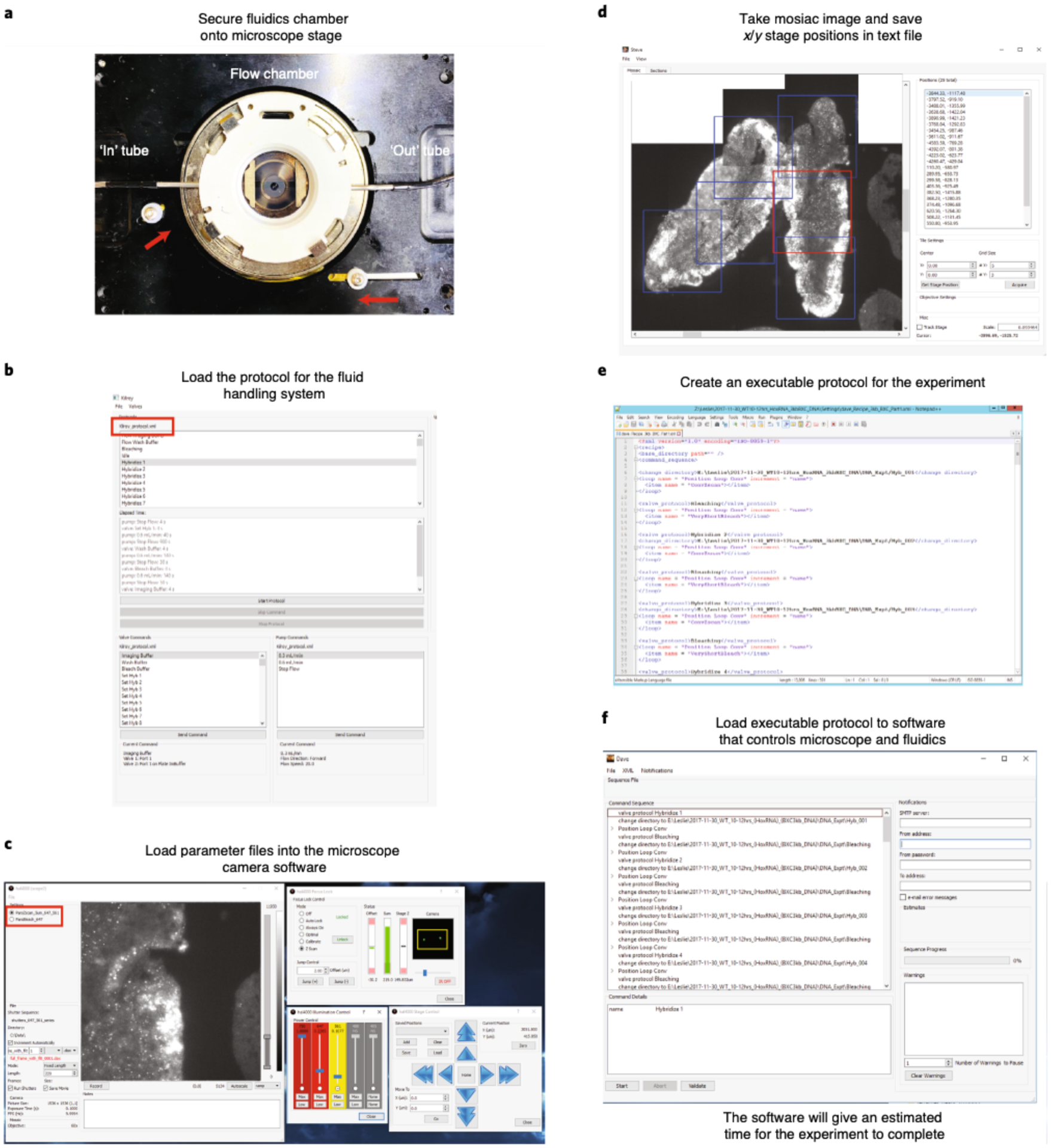
a, Tightly secure the flow chamber onto the microscope stage. Knobs on the stage have to be pushed in toward the flow chamber (shown with red arrows). Connect the ‘in’ and ‘out’ tubes (shown in white arrows) to the flow chamber. b, Open the fluidics system software and load the fluidics protocol. Examples of the fluidic Kilroy protocols are provided in the ‘Settings’ folder of the mini dataset (https://bit.ly/2S6eCjk), which are named Kilroy_15mins.xml and Kilroy_30mins.xml. The name of the loaded protocol is shown in the red box. Run Bleach Buffer through your sample to make sure that the flow is working properly. c, Open the microscope camera software and load the laser parameter files. Examples of the fluidic laser parameter files are provided in the ‘Settings’ folder of the mini dataset, which are named ParsBleach_647.xml and ParsZscan_5um_647_561.xml. The name of the parameters loaded is shown in the red box. Adjust the focus lock for your sample, which can be done in brightfield. The sample is in focus when you see two bright spots on the IR camera as shown inside the yellow box. In this example, we have already flown in the Cy3 fiducial oligo onto the cryosectioned Drosophila embryos that have been hybridized with RNA probes targeting 29 RNA transcripts. The 561-nm (Cy3) laser is used to visualize the fiducial signal for RNA primary probes. d, Using both the microscope camera software and the mosaic software, image a mosaic of your sample and manually record FOVs. Here we show a mosaic taken in the Cy3 channel. Example of a mosaic is in the ‘Mosaic’ folder of the mini dataset, which is named RNA_Brightfield_X.stv. Boxes represent the chosen FOVs where we purposely chose overlapping FOVs for downstream analysis. The red box indicates the currently selected FOV (which defaults to the last selected) The positions of these FOVs are saved and an example of these txt files (RNA_positions.txt and DNA_positions.txt) can be found in the ‘Settings’ folder of the mini dataset. e, The executable protocol can be copied and modified from a previous experiment. Here we show an example of a protocol. We have various examples of these protocols (‘dave.X.xml’) in the ‘Settings’ folder of the mini dataset. f, Open the Executable software and load in protocol from (e). Click ‘Validate’ to make sure that all parameter files are loaded onto their appropriate software and that the target folders, where data will be saved, exist. Afterward, click ‘Start’ to run your experiment.
