Figure 5.
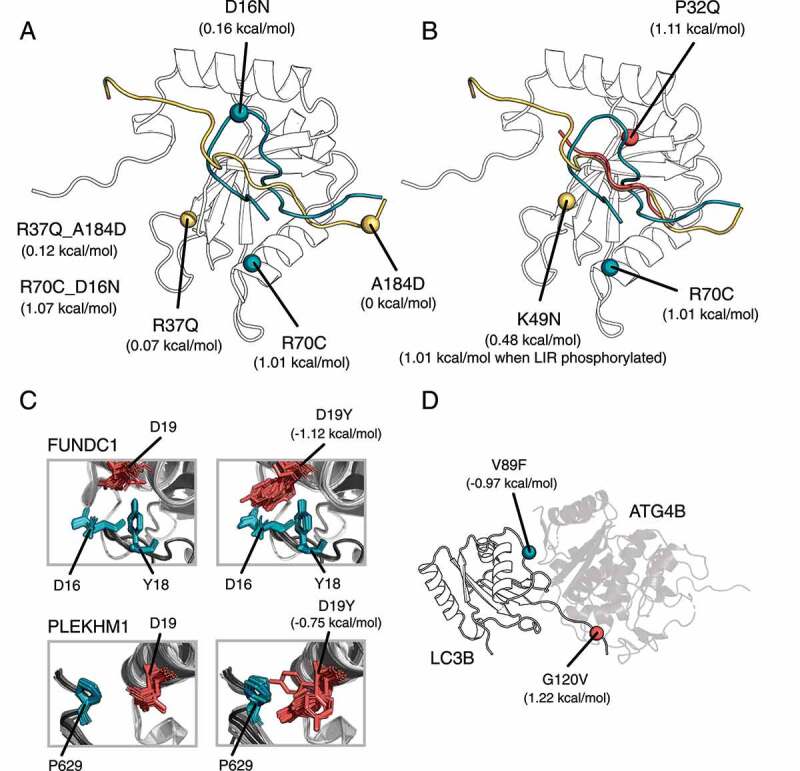
Changes in ΔΔG of binding for LC3B-LIR and LC3B-ATG4B complexes (Rosetta, talaris2014 energy function). (A) We illustrate the location and effect of the co-occurring mutations in the complex LC3B-OPTN (mutations and LIR in yellow), and in the complex LC3B-FUNDC1 (mutations and LIR in blue). Mutations of the first complex are neutral, whereas the mutations in LC3B-FUNDC1 complex destabilize the binding. (B) Mutations of LC3B can have a LIR-specific effect, as illustrated for P32Q in the complex LC3B-SQSTM1/p62 (mutation and LIR in red), K49N in the complex LC3B-OPTN (mutation and LIR in yellow) and for R70C in the complex LC3B-FUNDC1 (mutation and LIR in blue). (C) Mutations of LC3B could change specificity. We illustrate the effect of D19Y which may increase the binding affinity of PLEKHM1 and FUNDC1 to LC3B (D) V89F and G120V stabilizes and destabilizes the interaction between ATG4B and LC3B, respectively
