Figure 4.
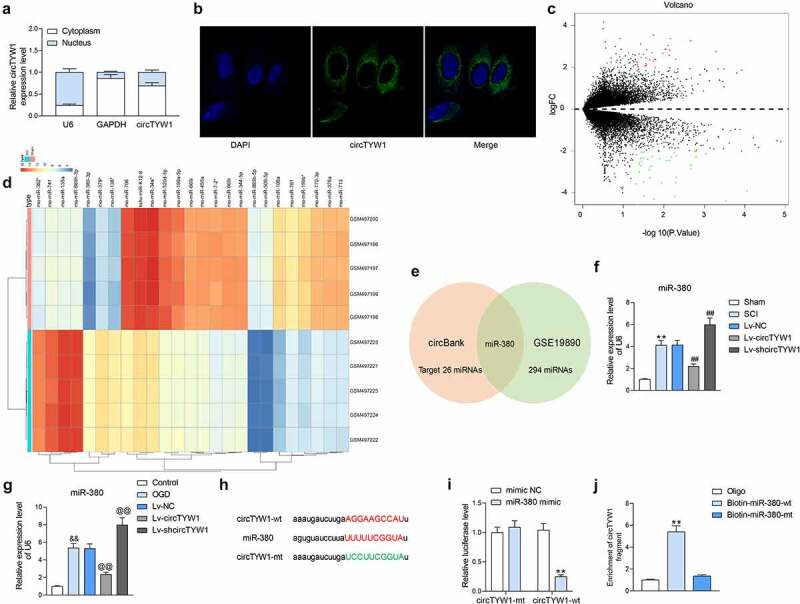
CircTYW1 is localized in the cytoplasm and binds to miR-380. A, subcellular localization of circTYW1 detected by fractionation and export assay; B, subcellular localization of circTYW1 detected by FISH experiments; C-D, GSE19890 microarray (comprised of spinal cord tissues from sham-operated 5 rats and 5 SCI rats) was performed for volcano plot (c) and heatmap (d) mapping; E, Venn diagram showing the number of miRNAs with binding relationship predicted by algorithm circbank and differentially expressed miRNAs in GSE19890 microarray; F, expression of miR-380 in rat spinal cord tissues on the 56th day after surgery by RT-qPCR (n = 6); G, expression of miR-380 in PC12 cells at 12 h after OGD treatment detected by RT-qPCR; H, binding sites of miR-380 to circTYW1; I, dual-luciferase assay determination of the binding relationship between miR-380 and circTYW1; J, biotin-labeled RNA pull down assay detection of the binding relationship between miR-380 and circTYW1. Data were presented as the mean ± SD. All experiments were repeated at least three times, and each experiment was performed in triplicate. One-way (panels F, G and J) or two-way ANOVA (panels A and I) with Tukey’s post-hoc test was used for statistical analysis. **p < 0.01 vs sham-operated rats, mimic NC or Oligo/miR-380-mt treatment; ##p < 0.01 vs SCI rats injected with Lv-NC; && p < 0.01 vs PC12 cells treated with normal condition; @@p < 0.01 vs PC12 cells transfected with Lv-NC and exposed to OGD
