Figure 1.
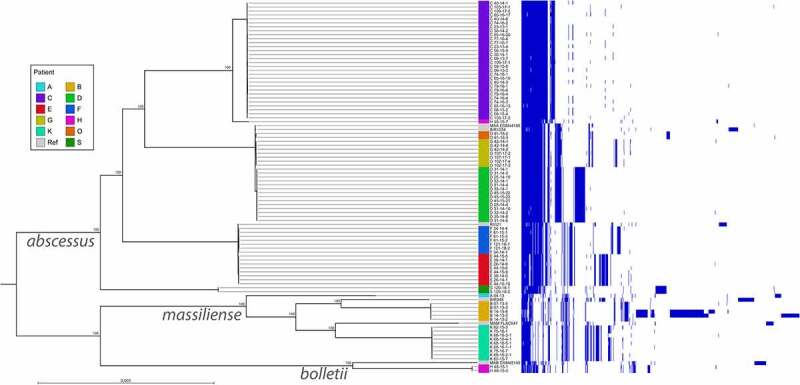
Phandango illustration showing a core genome-based Maximum likelihood tree corrected for recombination events of 88 M. abscessus isolates from 11 CF patients and the accessory genome. On the left side the Maximum likelihood tree is shown, on the right-side accessory gene presence in the respective isolates is indicated by blue lines. Patients providing samples are named by letters (A to S). Names of M. abscessus isolates are composed as follows: first letter stands for patient, first number for the sample number, second number for the year of isolation, last number for the colony number. Reference strains included in the tree were DSM 44196 (= ATCC 19977) for M. abscessus abscessus (accession NC_010397.1), CIP 108541 for M. abscessus bolletii (accession NZ_JRMF00000000.1) and FLAC047 for M. abscessus massiliense (accession NZ_CP021122.1). Representatives of the global patient-transmissible clusters [BIR 948 (accession ERS383065), RVI 21 (accession ERS244779), BIR 1034 (accession ERS383155)] described in [6] were included to identify global cluster strains present in the collection of CF isolates
