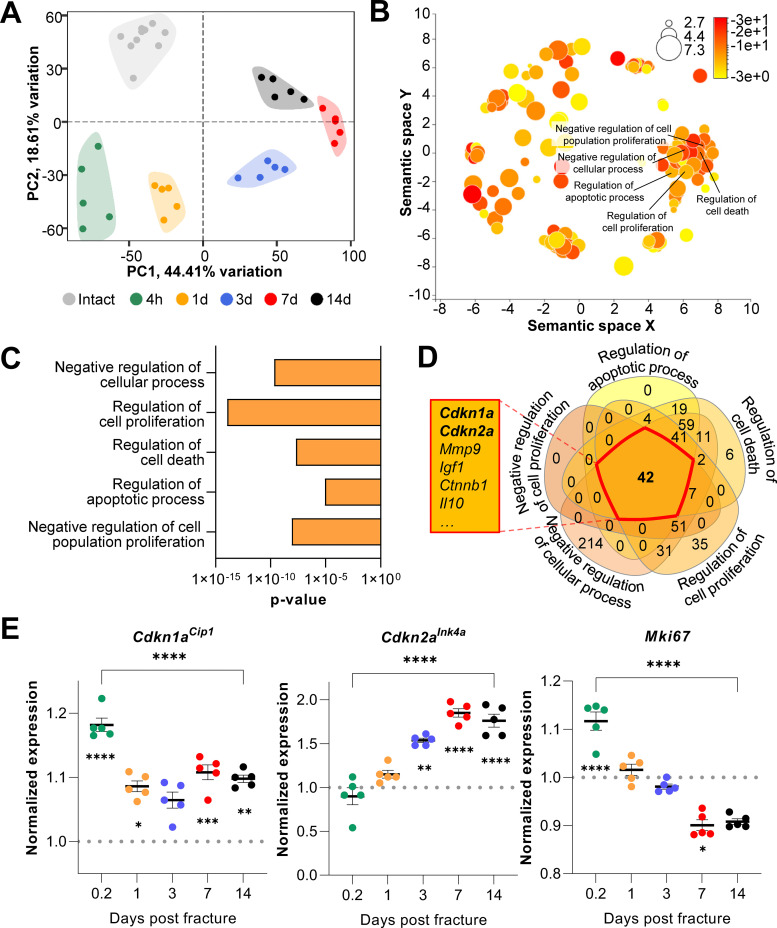
Figure 1. Cdkn1aCip1 and Cdkn2aInk4a are upregulated after the induction of femoral fractures in mice.
Transcriptome-wide changes during femoral fracture healing in mice were analyzed using publicly available mRNA-seq data (GSE152677 Coates et al., 2019). (A) Principal component analysis (PCA) illustrates the variance of gene expression profiles when comparing intact controls to fractured femora (0.2, 1, 3, 7, and 14 days post fracture; n = 5 per time point). (B) Cellular pathways affected by gene expression changes 14 days after fracture compared to intact controls were detected using gene ontology (REVIGO; Supek et al., 2011). (C) REVIGO revealed that pathways associated with cell proliferation and cell death (GO:0048523, GO:0042127, GO:0010941, GO:0042981, GO: 0008285) were significantly affected 14 days after a mid-diaphyseal stabilized femoral fracture. (D) When overlapping these pathways, 42 genes were found to be present in all gene sets, including Cdkn1aCip1 and Cdkn2aInk4a. (E) Normalized mRNA expression kinetics reveal an upregulation of Cdkn1aCip1 and Cdkn2aInk4a during fracture healing while Mki67 levels decreased. The dotted line represents the normalized baseline expression of intact control femora. The small asterisks indicate differences from the control. A-E: n = 35, all female. Mean ± SEM. One-way ANOVA; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.