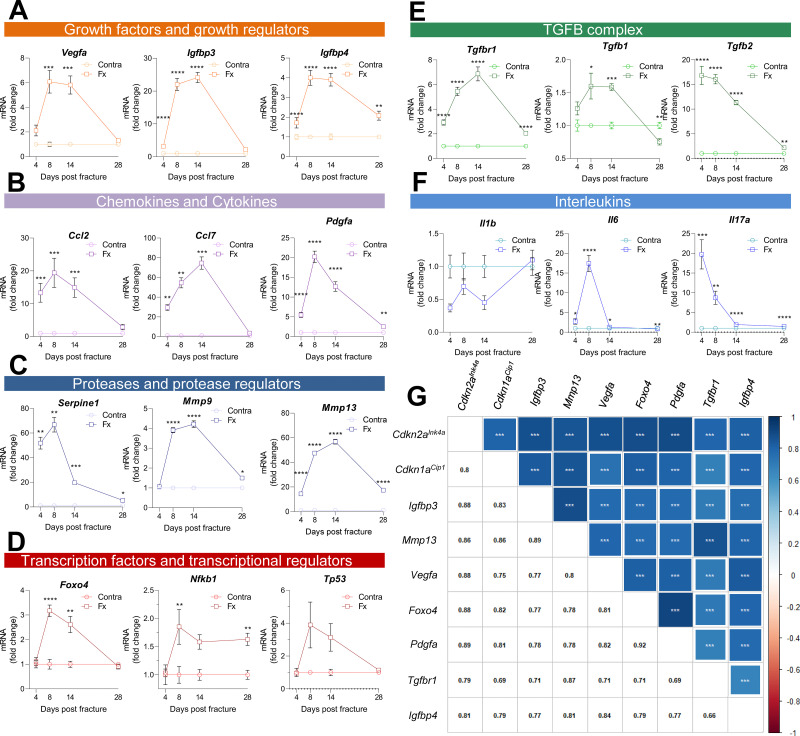
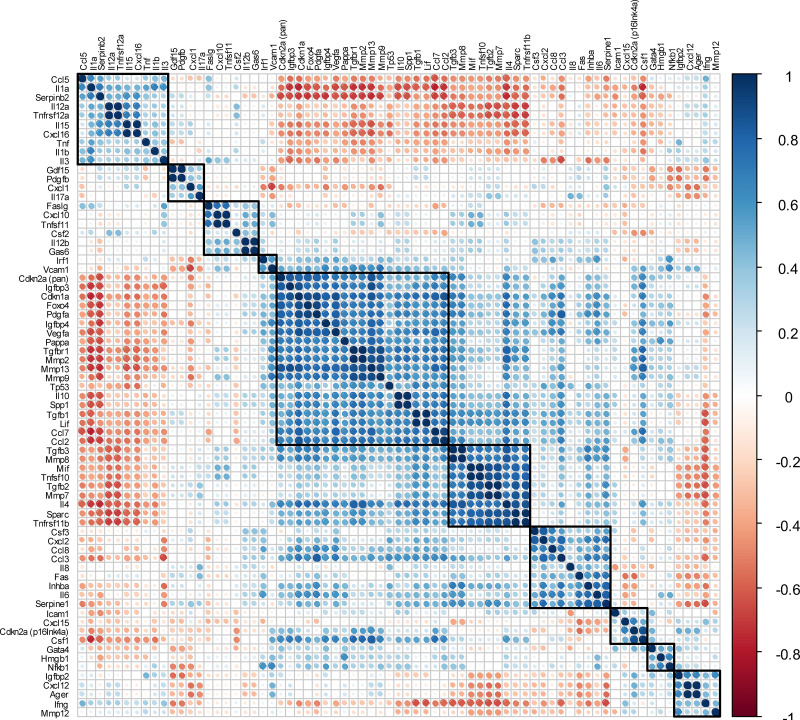
Figure 4. Multiple SASP factors show a marked increase in the healing callus, while distinct genes form similarly behaving clusters.
SASP-associated factors can be subdivided into functional subunits. (A) mRNA expression of the growth factors Vegfa, Igfbp3, and Igfbp4 substantially increased during the callus forming phase (days 2–15). (B) The chemokines Ccl2 and Pdgfa more profoundly rose in gene expression levels in the soft callus (days 2–7) but not in the hard callus (days 8–14) phase, while Ccl7 was more than 70-fold increased on day 14. (C) Expression of the protease Serpine1 was elevated in the soft callus phase, similar to Pdgfa, while Mmp9 and Mmp13 peaked on day 14. (D) The transcription factors Foxo4, Nfkb1, and Tp53 peaked on day 8, marking the beginning of hard callus formation. (E) TGFβ-associated genes displayed heterogeneous expression patterns. While Tgfbr1 peaked on day 14, Tgfb1 reached a plateau on days 8 and 14, and Tgfb2 gradually declined after an initial peak on day 4. (F) Among the interleukins, Il1b was negatively regulated in the beginning of the healing phase, while Il6 shortly peaked in the inflammatory and soft callus phase. Il17a showed a gradually decline, comparable to Tgfb2. (G) The largest cluster displaying similar gene expression patterns among all senescence-associated and SASP-gene markers included the key cell cycle regulators Cdkn2aInk4a and Cdkn1aCip1. The square size is proportional to the correlation coefficient, which is also depicted in the left bottom corner. Significant results were indicated with asterisks. A-G: n = 24 (n = 24 in Contra, n = 24 in Fx, 6 per time point per group), all male. Mean ± SEM. Multiple t-test (FDR); *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.