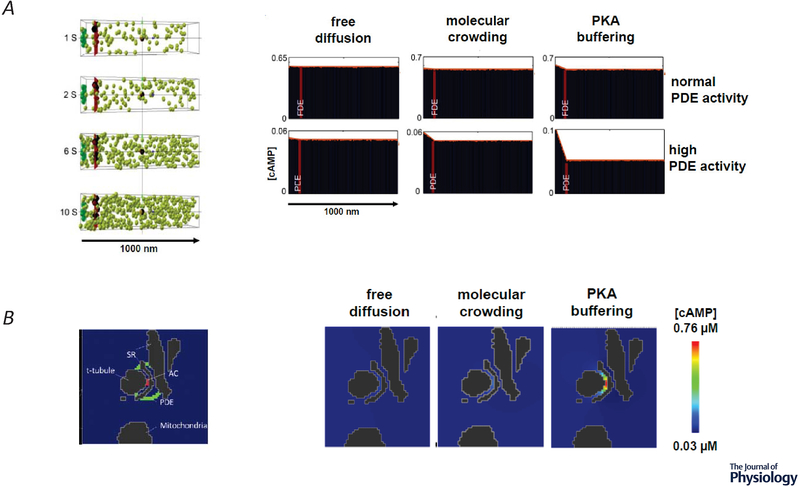
Figure 3.
A, Stochastic 3D simulation of cAMP diffusion. Left, four snapshots in time of the distribution of individual cAMP molecules (green spheres), generated by adenylyl cyclase (AC) activity (green box) in a caveolar membrane domain on the left, with cytosolic space to the right. Phosphodiesterase (PDE) molecules were placed in a plane (red bar) 100 nm from the plasma membrane to simulate a functional barrier. Right, average concentration of cAMP at various distances from the site of production when the number of PDE molecules and diffusion coefficient were varied. Normal PDE activity (415 μM), high PDE activity (41.5 mM). Free diffusion (200 μm2/s), molecular crowding (60 μm2/s), PKA buffering (10 μm2/s) B, 3D Continuum model of cAMP diffusion in an anatomically restricted space. Left, Cross section (1040 × 765 × 415 nm) through a 3D reconstruction of the dyadic space between the sarcoplasmic reticulum (SR) and t-tubule in an adult ventricular myocyte generated by cryo-transmission electron microscopy z-stacks. Location of AC activity is represented in red and PDE activity is represented in green. Right, Effect of diffusion coefficient on cAMP concentration in dyadic cleft in presence of normal PDE activity. Adapted from Yang et al. (2016).