Figure 1. Introduction to SSBP1 Protein Variants and SSBP1 Heterotetramers Tested.
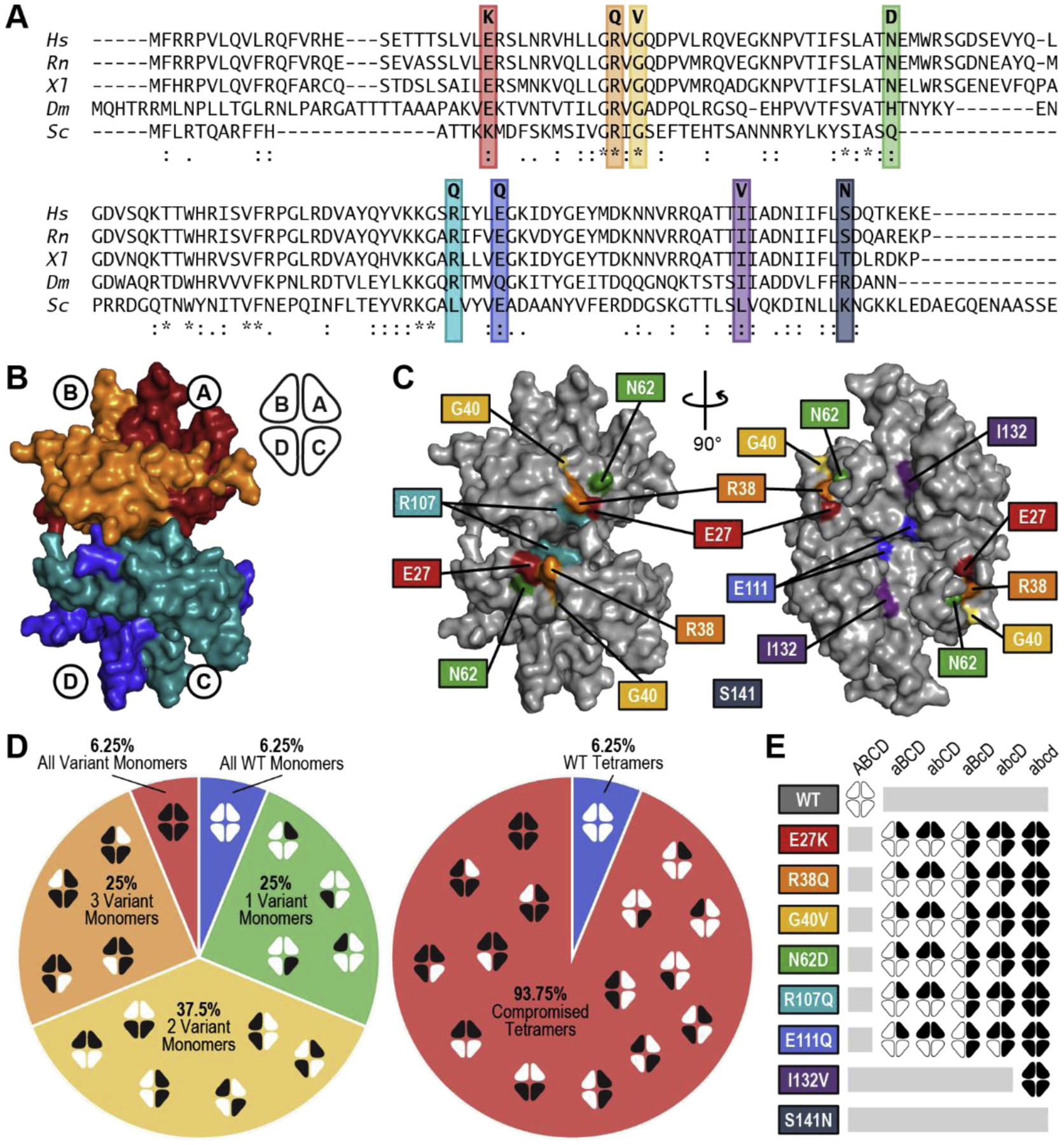
(A) Conservation of variant residues in SSBP1 from Homo sapiens (Hs) through Rattus norvegicus (Rn), Xenopus laevis (Xl), Drosophila melanogaster (Dm), and Saccharomyces cerevisiae (Sc). (*) perfect alignment; (:) strong similarity; (.) weak similarity. (B) Surface rendering of the SSBP1 tetramer (based on PBDID: 3ULL) with each monomer indicated, along with a simplified representation that will appear throughout this report. (C) Locations of the SSBP1 residues effected by disease mutations. Coordinates of S141 were not present in the reference crystal structure [9]. (D) Breakdown of all possible combinations of WT and variant monomers in SSBP1 tetramers in heterozygous patients, including ratios of all WT : single variant : double variant : triple variant : all variant tetramer systems (left) and WT : compromised tetramer systems (right). White indicates WT monomers; black indicates variant monomers. (E) Representation of 32 tetramer systems modeled in molecular dynamics simulations.
