Figure 7. DNA-Bound Structure Alignments and Dimethyl Phosphate Probing.
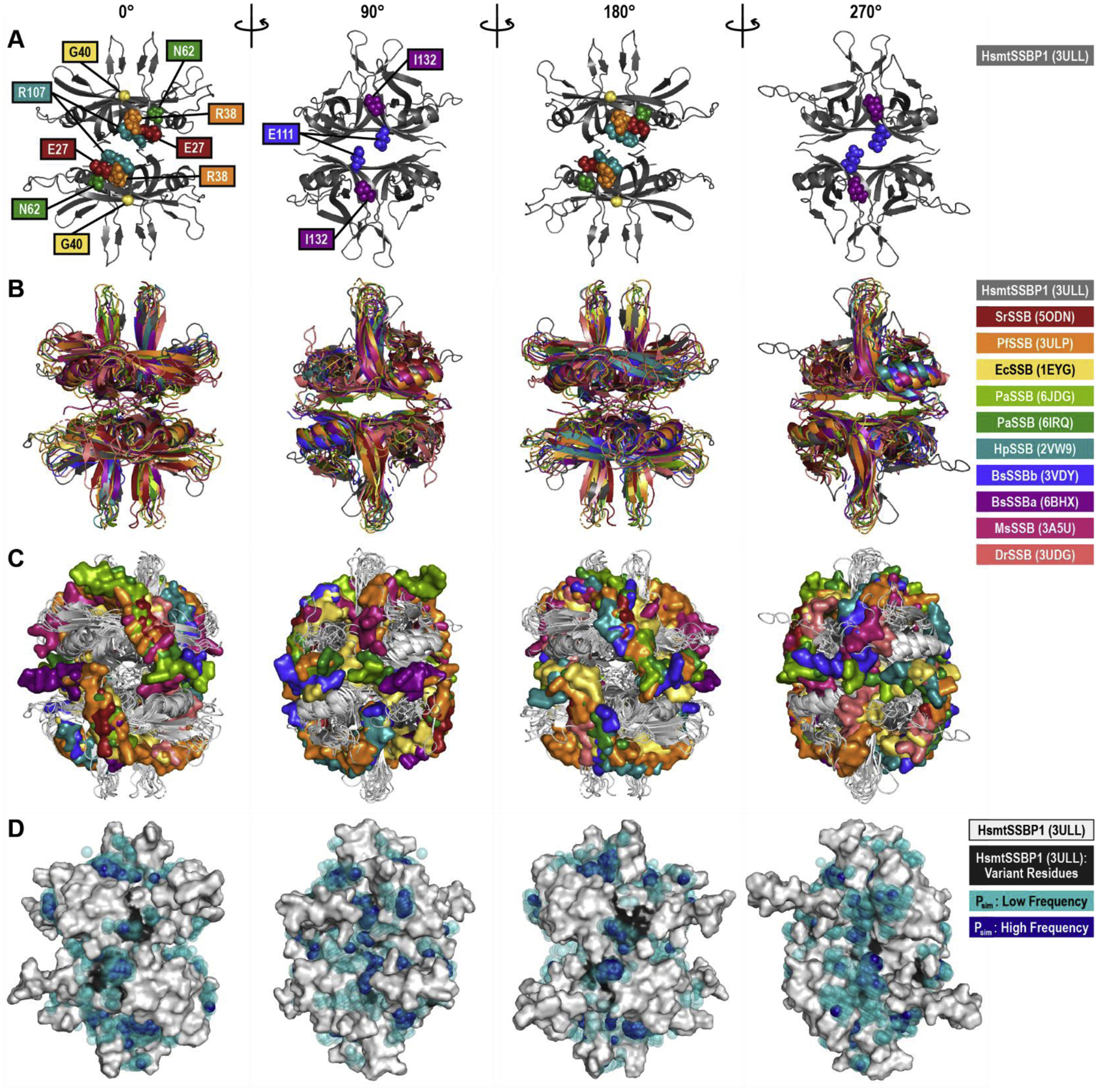
(A) The WT human mitochondrial (Hsmt) SSBP1 structure, based on PDB ID 3ULL and with missing loops modeled, is shown as a cartoon in four orientations, with residues linked to human disease indicated by spheres. Note that only the front-facing disease residues are shown in each orientation. This HsmtSSBP1 model was aligned with 10 other SSB proteins for which DNA-bound structures have been solved. This structural alignment is presented as cartoons of the proteins alone (B) and with the proteins rendered in white and each structure’s DNA rendered as surfaces (C). PDB IDs are listed in parentheses. (D) Predicted phosphate binding sites from molecular dynamics simulations are mapped onto a white surface rendering of HsmtSSBP1. Transparent teal spheres indicate low frequency dimethyl phosphate positions; dark blue opaque spheres indicate high frequency dimethyl phosphate positions. Black areas on the protein mark disease residue locations. Hs, Homo sapiens; Sr, Salinibacter ruber; Pf, Plasmodium falciparum; Ec, Escherichia coli; Pa, Pseudomonas aeruginosa; Hp, Helicobacter pylori; Bs, Bacillus subtilis; Ms, Mycolicibacterium smegmatis; Dr, Deinococcus radiodurans.
